New Insights into rice pyrimidine catabolic enzymes
- PMID: 36818891
- PMCID: PMC9930899
- DOI: 10.3389/fpls.2023.1079778
New Insights into rice pyrimidine catabolic enzymes
Abstract
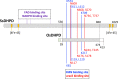
Introduction: Rice is a primary global food source, and its production is affected by abiotic stress, caused by climate change and other factors. Recently, the pyrimidine reductive catabolic pathway, catalyzed by dihydropyrimidine dehydrogenase (DHPD), dihydropyrimidinase (DHP) and β-ureidopropionase (β-UP), has emerged as a potential participant in the abiotic stress response of rice.
Methods: The rice enzymes were produced as recombinant proteins, and two were kinetically characterized. Rice dihydroorotate dehydrogenase (DHODH), an enzyme of pyrimidine biosynthesis often confused with DHPD, was also characterized. Salt-sensitive and salt-resistant rice seedlings were subjected to salt stress (24 h) and metabolites in leaves were determined by mass spectrometry.
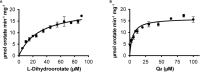
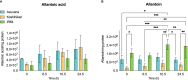
Results: The OsDHPD sequence was homologous to the C-terminal half of mammalian DHPD, conserving FMN and uracil binding sites, but lacked sites for Fe/S clusters, FAD, and NADPH. OsDHPD, truncated to eliminate the chloroplast targeting peptide, was soluble, but inactive. Database searches for polypeptides homologous to the N-terminal half of mammalian DHPD, that could act as co-reductants, were unsuccessful. OsDHODH exhibited kinetic parameters similar to those of other plant DHODHs. OsDHP, truncated to remove a signal sequence, exhibited a kcat/Km = 3.6 x 103 s-1M-1. Osb-UP exhibited a kcat/Km = 1.8 x 104 s-1M-1. Short-term salt exposure caused insignificant differences in the levels of the ureide intermediates dihydrouracil and ureidopropionate in leaves of salt-sensitive and salt-resistant plants. Allantoin, a ureide metabolite of purine catabolism, was found to be significantly higher in the resistant cultivar compared to one of the sensitive cultivars.
Discussion: OsDHP, the first plant enzyme to be characterized, showed low kinetic efficiency, but its activity may have been affected by truncation. Osb-UP exhibited kinetic parameters in the range of enzymes of secondary metabolism. Levels of two pathway metabolites were similar in sensitive and resistant cultivars and appeared to be unaffected by short-term salt exposure."
Keywords: Oryza sativa; abiotic stress; dihydroorotate dehydrogenase; dihydropyrimidinase; dihydropyrimidine dehydrogenase; plants; pyrimidine catabolism; ßureidopropionase.
Copyright © 2023 Lopez, Narvaez-Ortiz, Rincon-Benavides, Pulido, Fuentes Suarez and Zimmermann.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


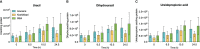
References
-
- Bower C. E., Holm-Hansen T. (1980). A salicylate–hypochlorite method for determining ammonia in seawater. Can. J. Fisheries Aquat. Sci. 37 (5), 794–798. doi: 10.1139/f80-106 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

