Advancements in Detection Approaches of Severe Acute Respiratory Syndrome Coronavirus 2
- PMID: 36818907
- PMCID: PMC9910375
- DOI: 10.21315/mjms2022.29.6.3
Advancements in Detection Approaches of Severe Acute Respiratory Syndrome Coronavirus 2
Abstract
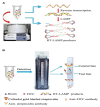
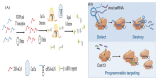
Diagnostic testing to identify individuals infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) plays a key role in selecting appropriate treatments, saving people's lives and preventing the global pandemic of COVID-19. By testing on a massive scale, some countries could successfully contain the disease spread. Since early viral detection may provide the best approach to curb the disease outbreak, the rapid and reliable detection of coronavirus (CoV) is therefore becoming increasingly important. Nucleic acid detection methods, especially real-time reverse transcription polymerase chain reaction (RT-PCR)-based assays are considered the gold standard for COVID-19 diagnostics. Some non-PCR-based molecular methods without thermocycler operation, such as isothermal nucleic acid amplification have been proved promising. Serologic immunoassays are also available. A variety of novel and improved methods based on biosensors, Clustered-Regularly Interspaced Short Palindromic Repeats (CRISPR) technology, lateral flow assay (LFA), microarray, aptamer etc. have also been developed. Several integrated, random-access, point-of-care (POC) molecular devices are rapidly emerging for quick and accurate detection of SARS-CoV-2 that can be used in the local hospitals and clinics. This review intends to summarize the currently available detection approaches of SARS-CoV-2, highlight gaps in existing diagnostic capacity, and propose potential solutions and thus may assist clinicians and researchers develop better technologies for rapid and authentic diagnosis of CoV infection.
Keywords: COVID-19; RT-qPCR; SARS-CoV-2; detection approach; nucleic acid.
© Penerbit Universiti Sains Malaysia, 2022.
Conflict of interest statement
Conflict of Interest None.
Figures

Similar articles
-
Recent advances and perspectives of nucleic acid detection for coronavirus.J Pharm Anal. 2020 Apr;10(2):97-101. doi: 10.1016/j.jpha.2020.02.010. Epub 2020 Mar 1. J Pharm Anal. 2020. PMID: 32292623 Free PMC article. Review.
-
Current state of diagnostic, screening and surveillance testing methods for COVID-19 from an analytical chemistry point of view.Microchem J. 2021 Aug;167:106305. doi: 10.1016/j.microc.2021.106305. Epub 2021 Apr 19. Microchem J. 2021. PMID: 33897053 Free PMC article.
-
[Recent advances in clustered regularly interspaced short palindromic repeats-based detection of severe acute respiratory syndrome coronavirus 2].Se Pu. 2022 Sep;40(9):773-781. doi: 10.3724/SP.J.1123.2022.08001. Se Pu. 2022. PMID: 36156623 Free PMC article. Review. Chinese.
-
Sensitive and rapid on-site detection of SARS-CoV-2 using a gold nanoparticle-based high-throughput platform coupled with CRISPR/Cas12-assisted RT-LAMP.Sens Actuators B Chem. 2021 Oct 15;345:130411. doi: 10.1016/j.snb.2021.130411. Epub 2021 Jul 6. Sens Actuators B Chem. 2021. PMID: 34248284 Free PMC article.
-
CLEVER assay: A visual and rapid RNA extraction-free detection of SARS-CoV-2 based on CRISPR-Cas integrated RT-LAMP technology.J Appl Microbiol. 2022 Aug;133(2):410-421. doi: 10.1111/jam.15571. Epub 2022 Apr 18. J Appl Microbiol. 2022. PMID: 35396760 Free PMC article.
Cited by
-
Development and clinical application of loop-mediated isothermal amplification combined with lateral flow assay for rapid diagnosis of SARS-CoV-2.BMC Infect Dis. 2024 Jan 15;24(1):81. doi: 10.1186/s12879-023-08924-3. BMC Infect Dis. 2024. PMID: 38225546 Free PMC article.
-
Real-time PCR assays that detect genes for botulinum neurotoxin A-G subtypes.Front Microbiol. 2024 May 30;15:1382056. doi: 10.3389/fmicb.2024.1382056. eCollection 2024. Front Microbiol. 2024. PMID: 38873139 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
