Remote visualization of large-scale genomic alignments for collaborative clinical research and diagnosis of rare diseases
- PMID: 36819661
- PMCID: PMC9932977
- DOI: 10.1016/j.xgen.2022.100246
Remote visualization of large-scale genomic alignments for collaborative clinical research and diagnosis of rare diseases
Abstract
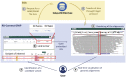
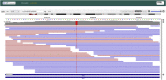
The Solve-RD project objectives include solving undiagnosed rare diseases (RD) through collaborative research on shared genome-phenome datasets. The RD-Connect Genome-Phenome Analysis Platform (GPAP), for data collation and analysis, and the European Genome-Phenome Archive (EGA), for file storage, are two key components of the Solve-RD infrastructure. Clinical researchers can identify candidate genetic variants within the RD-Connect GPAP and, thanks to the developments presented here as part of joint ELIXIR activities, are able to remotely visualize the corresponding alignments stored at the EGA. The Global Alliance for Genomics and Health (GA4GH) htsget streaming application programming interface (API) is used to retrieve alignment slices, which are rendered by an integrated genome viewer (IGV) instance embedded in the GPAP. As a result, it is no longer necessary for over 11,000 datasets to download large alignment files to visualize them locally. This work highlights the advantages, from both the user and infrastructure perspectives, of implementing interoperability standards for establishing federated genomics data networks.
Keywords: data sharing; data visualization; diagnosis; exome analysis; federated infrastructures; genome analysis; rare diseases; remote data access; standards.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Farwell K.D., Shahmirzadi L., El-Khechen D., Powis Z., Chao E.C., Tippin Davis B., Baxter R.M., Zeng W., Mroske C., Parra M.C., et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 2015;17:578–586. doi: 10.1038/gim.2014.154. - DOI - PubMed
-
- Stark Z., Tan T.Y., Chong B., Brett G.R., Yap P., Walsh M., Yeung A., Peters H., Mordaunt D., Cowie S., et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet. Med. 2016;18:1090–1096. doi: 10.1038/gim.2016.1. - DOI - PubMed
-
- Wright C.F., Fitzgerald T.W., Jones W.D., Clayton S., McRae J.F., van Kogelenberg M., King D.A., Ambridge K., Barrett D.M., Bayzetinova T., et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385:1305–1314. doi: 10.1016/S0140-6736(14)61705-0. - DOI - PMC - PubMed
-
- Zurek B., Ellwanger K., Vissers L.E.L.M., Schüle R., Synofzik M., Töpf A., de Voer R.M., Laurie S., Matalonga L., Gilissen C., et al. Solve-RD consortium Solve-RD: systematic pan-European data sharing and collaborative analysis to solve rare diseases. Eur. J. Hum. Genet. 2021;29:1325–1331. doi: 10.1038/s41431-021-00859-0. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

