Reactive gene curation to support interpretation and reporting of a clinical genome test for rare disease: Experience from over 1,000 cases
- PMID: 36819666
- PMCID: PMC9932986
- DOI: 10.1016/j.xgen.2023.100258
Reactive gene curation to support interpretation and reporting of a clinical genome test for rare disease: Experience from over 1,000 cases
Abstract
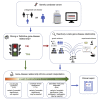
Current standards in clinical genetics recognize the need to establish the validity of gene-disease relationships as a first step in the interpretation of sequence variants. We describe our experience incorporating the ClinGen Gene-Disease Clinical Validity framework in our interpretation and reporting workflow for a clinical genome sequencing (cGS) test for individuals with rare and undiagnosed genetic diseases. This "reactive" gene curation is completed upon identification of candidate variants during active case analysis and within the test turn-around time by focusing on the most impactful evidence and taking advantage of the broad applicability of the framework to cover a wide range of disease areas. We demonstrate that reactive gene curation can be successfully implemented in support of cGS in a clinical laboratory environment, enabling robust clinical decision making and allowing all variants to be fully and appropriately considered and their clinical significance confidently interpreted.
Keywords: ClinGen; GDR; RUGD; candidate gene; disease gene; gene curation; gene discovery; gene-disease relationship; genome sequencing; rare disease.
© 2023 The Author(s).
Conflict of interest statement
All authors are employees and shareholders of Illumina, Inc.
Figures



References
-
- Lionel A.C., Costain G., Monfared N., Walker S., Reuter M.S., Hosseini S.M., Thiruvahindrapuram B., Merico D., Jobling R., Nalpathamkalam T., et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet. Med. 2018;20:435–443. doi: 10.1038/gim.2017.119. - DOI - PMC - PubMed
-
- Scocchia A., Wigby K.M., Masser-Frye D., Del Campo M., Galarreta C.I., Thorpe E., McEachern J., Robinson K., Gross A., et al. ICSL Interpretation and Reporting Team Clinical whole genome sequencing as a first-tier test at a resource-limited dysmorphology clinic in Mexico. NPJ Genom. Med. 2019;4:5. doi: 10.1038/s41525-018-0076-1. - DOI - PMC - PubMed
-
- Bertoli-Avella A.M., Beetz C., Ameziane N., Rocha M.E., Guatibonza P., Pereira C., Calvo M., Herrera-Ordonez N., Segura-Castel M., Diego-Alvarez D., et al. Successful application of genome sequencing in a diagnostic setting: 1007 index cases from a clinically heterogeneous cohort. Eur. J. Hum. Genet. 2021;29:141–153. doi: 10.1038/s41431-020-00713-9. - DOI - PMC - PubMed
-
- NICUSeq Study Group. Krantz I.D., Medne L., Weatherly J.M., Wild K.T., Biswas S., Devkota B., Hartman T., Brunelli L., Fishler K.P., et al. Effect of whole-genome sequencing on the clinical management of acutely ill infants with suspected genetic disease: a randomized clinical trial. JAMA Pediatr. 2021;175:1218–1226. doi: 10.1001/jamapediatrics.2021.3496. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

