The Michigan Genomics Initiative: A biobank linking genotypes and electronic clinical records in Michigan Medicine patients
- PMID: 36819667
- PMCID: PMC9932985
- DOI: 10.1016/j.xgen.2023.100257
The Michigan Genomics Initiative: A biobank linking genotypes and electronic clinical records in Michigan Medicine patients
Abstract
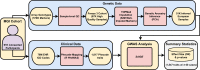
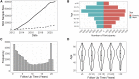
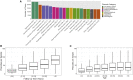
Biobanks of linked clinical patient histories and biological samples are an efficient strategy to generate large cohorts for modern genetics research. Biobank recruitment varies by factors such as geographic catchment and sampling strategy, which affect biobank demographics and research utility. Here, we describe the Michigan Genomics Initiative (MGI), a single-health-system biobank currently consisting of >91,000 participants recruited primarily during surgical encounters at Michigan Medicine. The surgical enrollment results in a biobank enriched for many diseases and ideally suited for a disease genetics cohort. Compared with the much larger population-based UK Biobank, MGI has higher prevalence for nearly all diagnosis-code-based phenotypes and larger absolute case counts for many phenotypes. Genome-wide association study (GWAS) results replicate known findings, thereby validating the genetic and clinical data. Our results illustrate that opportunistic biobank sampling within single health systems provides a unique and complementary resource for exploring the genetics of complex diseases.
© 2023 The Authors.
Conflict of interest statement
G.R.A. and A.P. work for Regeneron Pharmaceuticals. C.J.W. took a position at Regeneron Pharmaceuticals after the initial submission of this manuscript.
Figures



References
-
- Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. - DOI - PMC - PubMed
-
- Beesley L.J., Salvatore M., Fritsche L.G., Pandit A., Rao A., Brummett C., Willer C.J., Lisabeth L.D., Mukherjee B. The emerging landscape of health research based on biobanks linked to electronic health records: existing resources, statistical challenges, and potential opportunities. Stat. Med. 2020;39:773–800. doi: 10.1002/sim.8445. - DOI - PMC - PubMed
-
- Denny J.C., Ritchie M.D., Basford M.A., Pulley J.M., Bastarache L., Brown-Gentry K., Wang D., Masys D.R., Roden D.M., Crawford D.C. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. - DOI - PMC - PubMed
-
- Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., et al. Genome-wide genetic data on ∼500,000 UK Biobank participants. bioRxiv. 2017 doi: 10.1101/166298. Preprint at. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

