A searchable image resource of Drosophila GAL4 driver expression patterns with single neuron resolution
- PMID: 36820523
- PMCID: PMC10030108
- DOI: 10.7554/eLife.80660
A searchable image resource of Drosophila GAL4 driver expression patterns with single neuron resolution
Abstract
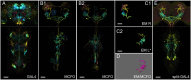
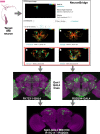
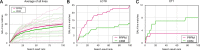
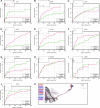
Precise, repeatable genetic access to specific neurons via GAL4/UAS and related methods is a key advantage of Drosophila neuroscience. Neuronal targeting is typically documented using light microscopy of full GAL4 expression patterns, which generally lack the single-cell resolution required for reliable cell type identification. Here, we use stochastic GAL4 labeling with the MultiColor FlpOut approach to generate cellular resolution confocal images at large scale. We are releasing aligned images of 74,000 such adult central nervous systems. An anticipated use of this resource is to bridge the gap between neurons identified by electron or light microscopy. Identifying individual neurons that make up each GAL4 expression pattern improves the prediction of split-GAL4 combinations targeting particular neurons. To this end, we have made the images searchable on the NeuronBridge website. We demonstrate the potential of NeuronBridge to rapidly and effectively identify neuron matches based on morphology across imaging modalities and datasets.
Keywords: D. melanogaster; GAL4; MultiColor FlpOut; NeuronBridge; neuron search; neuroscience; split-GAL4.
© 2023, Meissner et al.
Conflict of interest statement
GM, AN, ZD, GD, KF, TG, JH, YH, NI, JJ, LJ, RJ, KL, BM, BY, CZ, JC, CG, HO, KR, RS, YA, GC, BD, EE, JG, MI, DK, WK, LM, RM, SN, GR, GS, TW, OM No competing interests declared
Figures


Update of
References
-
- Aso Y, Sitaraman D, Ichinose T, Kaun KR, Vogt K, Belliart-Guérin G, Plaçais PY, Robie AA, Yamagata N, Schnaitmann C, Rowell WJ, Johnston RM, Ngo TTB, Chen N, Korff W, Nitabach MN, Heberlein U, Preat T, Branson KM, Tanimoto H, Rubin GM. Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. eLife. 2014b;3:e04580. doi: 10.7554/eLife.04580. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

