Upper respiratory tract microbiota dynamics following COVID-19 in adults
- PMID: 36820832
- PMCID: PMC9997743
- DOI: 10.1099/mgen.0.000957
Upper respiratory tract microbiota dynamics following COVID-19 in adults
Abstract
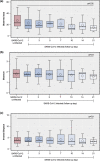
To date, little is known about the effect of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus responsible for the coronavirus disease 2019 (COVID-19) pandemic, on the upper respiratory tract (URT) microbiota over time. To fill this knowledge gap, we used 16S ribosomal RNA gene sequencing to characterize the URT microbiota in 48 adults, including (1) 24 participants with mild-to-moderate COVID-19 who had serial mid-turbinate swabs collected up to 21 days after enrolment and (2) 24 asymptomatic, uninfected controls who had mid-turbinate swabs collected at enrolment only. To compare the URT microbiota between groups in a comprehensive manner, different types of statistical analyses that are frequently employed in microbial ecology were used, including ⍺-diversity, β-diversity and differential abundance analyses. Final statistical models included age, sex and the presence of at least one comorbidity as covariates. The median age of all participants was 34.00 (interquartile range=28.75-46.50) years. In comparison to samples from controls, those from participants with COVID-19 had a lower observed species index at day 21 (linear regression coefficient=-13.30; 95 % CI=-21.72 to -4.88; q=0.02). In addition, the Jaccard index was significantly different between samples from participants with COVID-19 and those from controls at all study time points (PERMANOVA q<0.05 for all comparisons). The abundance of three amplicon sequence variants (ASVs) (one Corynebacterium ASV, Frederiksenia canicola, and one Lactobacillus ASV) were decreased in samples from participants with COVID-19 at all seven study time points, whereas the abundance of one ASV (from the family Neisseriaceae) was increased in samples from participants with COVID-19 at five (71.43 %) of the seven study time points. Our results suggest that mild-to-moderate COVID-19 can lead to alterations of the URT microbiota that persist for several weeks after the initial infection.
Keywords: COVID-19; SARS-CoV-2; airway; coronavirus; microbiota; nasal; respiratory.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures





References
-
- Rosas-Salazar C, Shilts MH, Tovchigrechko A, Chappell JD, Larkin EK, et al. Nasopharyngeal microbiome in respiratory syncytial virus resembles profile associated with increased childhood Asthma risk. Am J Respir Crit Care Med. 2016;193:1180–1183. doi: 10.1164/rccm.201512-2350LE. - DOI - PMC - PubMed
-
- Rosas-Salazar C, Shilts MH, Tovchigrechko A, Schobel S, Chappell JD, et al. Differences in the nasopharyngeal microbiome during acute respiratory tract infection with human rhinovirus and respiratory syncytial virus in infancy. J Infect Dis. 2016;214:1924–1928. doi: 10.1093/infdis/jiw456. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

