Reversible epigenetic alterations mediate PSMA expression heterogeneity in advanced metastatic prostate cancer
- PMID: 36821396
- PMCID: PMC10132157
- DOI: 10.1172/jci.insight.162907
Reversible epigenetic alterations mediate PSMA expression heterogeneity in advanced metastatic prostate cancer
Abstract
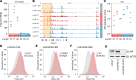
Prostate-specific membrane antigen (PSMA) is an important cell surface target in prostate cancer. There are limited data on the heterogeneity of PSMA tissue expression in metastatic castration-resistant prostate cancer (mCRPC). Furthermore, the mechanisms regulating PSMA expression (encoded by the FOLH1 gene) are not well understood. Here, we demonstrate that PSMA expression is heterogeneous across different metastatic sites and molecular subtypes of mCRPC. In a rapid autopsy cohort in which multiple metastatic sites per patient were sampled, we found that 13 of 52 (25%) cases had no detectable PSMA and 23 of 52 (44%) cases showed heterogeneous PSMA expression across individual metastases, with 33 (63%) cases harboring at least 1 PSMA-negative site. PSMA-negative tumors displayed distinct transcriptional profiles with expression of druggable targets such as MUC1. Loss of PSMA was associated with epigenetic changes of the FOLH1 locus, including gain of CpG methylation and loss of histone 3 lysine 27 (H3K27) acetylation. Treatment with histone deacetylase (HDAC) inhibitors reversed this epigenetic repression and restored PSMA expression in vitro and in vivo. Collectively, these data provide insights into the expression patterns and regulation of PSMA in mCRPC and suggest that epigenetic therapies - in particular, HDAC inhibitors - can be used to augment PSMA levels.
Keywords: Drug therapy; Epigenetics; Oncology; Prostate cancer.
Conflict of interest statement
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

