Ultrasensitive and Specific Identification of Monkeypox Virus Congo Basin and West African Strains Using a CRISPR/Cas12b-Based Platform
- PMID: 36821485
- PMCID: PMC10100855
- DOI: 10.1128/spectrum.04035-22
Ultrasensitive and Specific Identification of Monkeypox Virus Congo Basin and West African Strains Using a CRISPR/Cas12b-Based Platform
Abstract
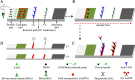
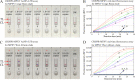
Human monkeypox (MPX) is a severe and reemerging infectious disease caused by monkeypox virus (MPXV) and forms two distinct lineages, including Congo Basin and West African clades. Due to the absence of specific vaccines and antiviral drugs, developing a point-of-care (POC) testing system to identify MPXV is critical for preventing and controlling MPX transmission. Here, a CRISPR/Cas12b diagnostic platform was integrated with loop-mediated isothermal amplification (LAMP) to devise a novel CRISPR-MPXV approach for ultrasensitive, highly specific, rapid, and simple detection of MPXV Congo Basin and West African strains, and the detection results were interpreted with real-time fluorescence and a gold nanoparticle-based lateral flow biosensor (AuNP-LFB). The optimal detection process, including genomic DNA extraction (15 min), LAMP preamplification (35 min at 66°C), CRISPR/Cas12b-based detection (5 min at 45°C), and AuNP-LFB readout (~2 min), can be completed within 60 min without expensive instruments. Our assay has a limit of detection of 10 copies per test and produces no cross-reaction with any other types of pathogens. Hence, our CRISPR-MPXV assay exhibited considerable potential for POC testing for identifying and distinguishing MPXV Congo Basin and West African strains, especially in regions with resource shortages. IMPORTANCE Monkeypox (MPX), a reemerging zoonotic disease caused by monkeypox virus (MPXV), causes a smallpox-like disease in humans. Early diagnosis is critical to prevent MPX epidemics. Here, CRISPR/Cas12b was integrated with LAMP amplification to devise a novel CRISPR-MPXV approach to achieve highly specific, ultrasensitive, rapid, and visual identification of MPXV Congo Basin and West African strains.
Keywords: CRISPR/Cas12b; gold nanoparticle-based lateral flow biosensor; loop-mediated isothermal amplification; monkeypox; point-of-care testing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Loop-mediated isothermal amplification combined with lateral flow biosensor for rapid and sensitive detection of monkeypox virus.Front Public Health. 2023 Mar 24;11:1132896. doi: 10.3389/fpubh.2023.1132896. eCollection 2023. Front Public Health. 2023. PMID: 37033067 Free PMC article.
-
Loop-mediated isothermal amplification coupled with nanoparticle-based lateral flow biosensor for monkeypox virus detection.Talanta. 2024 Mar 1;269:125502. doi: 10.1016/j.talanta.2023.125502. Epub 2023 Dec 1. Talanta. 2024. PMID: 38070288
-
Loop-mediated isothermal amplification-based diagnostic assay for monkeypox virus infections.J Med Virol. 2009 Jun;81(6):1102-8. doi: 10.1002/jmv.21494. J Med Virol. 2009. PMID: 19382264
-
Monkeypox virus: a re-emergent threat to humans.Virol Sin. 2022 Aug;37(4):477-482. doi: 10.1016/j.virs.2022.07.006. Epub 2022 Jul 9. Virol Sin. 2022. PMID: 35820590 Free PMC article. Review.
-
Strategy of developing nucleic acid-based universal monkeypox vaccine candidates.Front Immunol. 2022 Oct 27;13:1050309. doi: 10.3389/fimmu.2022.1050309. eCollection 2022. Front Immunol. 2022. PMID: 36389680 Free PMC article. Review.
Cited by
-
CRISPR-based strategies for sample-to-answer monkeypox detection: current status and emerging opportunities.Nanotechnology. 2024 Nov 4;36(4):042001. doi: 10.1088/1361-6528/ad892b. Nanotechnology. 2024. PMID: 39433062 Free PMC article. Review.
-
Mpox disease, diagnosis, and point of care platforms.Bioeng Transl Med. 2025 Jan 2;10(3):e10733. doi: 10.1002/btm2.10733. eCollection 2025 May. Bioeng Transl Med. 2025. PMID: 40385539 Free PMC article. Review.
-
A novel single-tube LAMP-CRISPR/Cas12b method for rapid and visual detection of zoonotic Toxoplasma gondii in the environment.Infect Dis Poverty. 2024 Dec 10;13(1):94. doi: 10.1186/s40249-024-01266-5. Infect Dis Poverty. 2024. PMID: 39654027 Free PMC article.
-
Rapid visual detection of Monkeypox virus by one-step LAMP-CRISPR/Cas12b assay.Virol J. 2025 May 20;22(1):151. doi: 10.1186/s12985-025-02780-0. Virol J. 2025. PMID: 40394594 Free PMC article.
-
Detection of monkeypox virus using helicase dependent amplification and recombinase polymerase amplification combined with lateral flow test.Virol J. 2023 Nov 23;20(1):274. doi: 10.1186/s12985-023-02223-8. Virol J. 2023. PMID: 37996921 Free PMC article.
References
-
- Vivancos R, Anderson C, Blomquist P, Balasegaram S, Bell A, Bishop L, Brown CS, Chow Y, Edeghere O, Florence I, Logan S, Manley P, Crowe W, McAuley A, Shankar AG, Mora-Peris B, Paranthaman K, Prochazka M, Ryan C, Simons D, Vipond R, Byers C, Watkins NA, Welfare W, Whittaker E, Dewsnap C, Wilson A, Young Y, Chand M, Riley S, Hopkins S, UKHSA Monkeypox Incident Management Team . 2022. Community transmission of monkeypox in the United Kingdom, April to May 2022. Euro Surveill 27:2200422. doi: 10.2807/1560-7917.ES.2022.27.22.2200422. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

