Defining and targeting tumor-associated macrophages in malignant mesothelioma
- PMID: 36821580
- PMCID: PMC9992826
- DOI: 10.1073/pnas.2210836120
Defining and targeting tumor-associated macrophages in malignant mesothelioma
Abstract
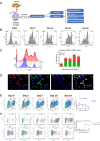
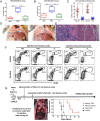
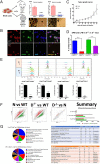
Defining the ontogeny of tumor-associated macrophages (TAM) is important to develop therapeutic targets for mesothelioma. We identified two distinct macrophage populations in mouse peritoneal and pleural cavities, the monocyte-derived, small peritoneal/pleural macrophages (SPM), and the tissue-resident large peritoneal/pleural macrophages (LPM). SPM rapidly increased in tumor microenvironment after tumor challenge and contributed to the vast majority of M2-like TAM. The selective depletion of M2-like TAM by conditional deletion of the Dicer1 gene in myeloid cells (D-/-) promoted tumor rejection. Sorted SPM M2-like TAM initiated tumorigenesis in vivo and in vitro, confirming their capacity to support tumor development. The transcriptomic and single-cell RNA sequencing analysis demonstrated that both SPM and LPM contributed to the tumor microenvironment by promoting the IL-2-STAT5 signaling pathway, inflammation, and epithelial-mesenchymal transition. However, while SPM preferentially activated the KRAS and TNF-α/NFkB signaling pathways, LPM activated the IFN-γ response. The importance of LPM in the immune response was confirmed by depleting LPM with intrapleural clodronate liposomes, which abrogated the antitumoral memory immunity. SPM gene signature could be identified in pleural effusion and tumor from patients with untreated mesothelioma. Five genes, TREM2, STAB1, LAIR1, GPNMB, and MARCO, could potentially be specific therapeutic targets. Accordingly, Trem2 gene deletion led to reduced SPM M2-like TAM with compensatory increase in LPM and slower tumor growth. Overall, these experiments demonstrate that SPM M2-like TAM play a key role in mesothelioma development, while LPM more specifically contribute to the immune response. Therefore, selective targeting of monocyte-derived TAM may enhance antitumor immunity through compensatory expansion of tissue-resident TAM.
Keywords: cancer; gene signature; mesothelioma; tumor microenvironment; tumor-associated macrophages.
Conflict of interest statement
The authors have organizational affiliations to disclose. M. De Perrot received personal fees outside of this work from Bayer (speaking fees for work related to pulmonary hypertension), Merck (speaking fees for work related to lung cancer), Janssen (consulting fees for work related to pulmonary hypertension), Roche (consulting fees for work related to lung cancer), BMS (consulting fees for work related to lung cancer), and Astra-Zeneca (Ad board for work related to lung cancer).
Figures





References
-
- Courtiol P., et al. , Deep learning-based classification of mesothelioma improves prediction of patient outcome. Nat. Med. 25, 1519–1525 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

