MicroED in drug discovery
- PMID: 36821888
- PMCID: PMC10023408
- DOI: 10.1016/j.sbi.2023.102549
MicroED in drug discovery
Abstract
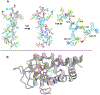
The cryo-electron microscopy (cryo-EM) method microcrystal electron diffraction (MicroED) was initially described in 2013 and has recently gained attention as an emerging technique for research in drug discovery. As compared to other methods in structural biology, MicroED provides many advantages deriving from the use of nanocrystalline material for the investigations. Here, we review the recent advancements in the field of MicroED and show important examples of small molecule, peptide and protein structures that has contributed to the current development of this method as an important tool for drug discovery.
Keywords: Cryo-EM; MicroED; Nanocrystals; Protein-ligand structures; Small molecule structures.
Copyright © 2023 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
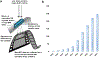


References
-
- van Genderen E, Clabbers MT, Das PP, Stewart A, Nederlof I, Barentsen KC, Portillo Q, Pannu NS, Nicolopoulos S, Gruene T, Abrahams JP: Ab initio structure determination of nanocrystals of organic pharmaceutical compounds by electron diffraction at room temperature using a Timepix quantum area direct electron detector. Acta Crystallogr A 2016, 72:236–242. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

