Review: A silent concert in developing plants: Dynamic assembly of cullin-RING ubiquitin ligases
- PMID: 36822503
- PMCID: PMC10065934
- DOI: 10.1016/j.plantsci.2023.111662
Review: A silent concert in developing plants: Dynamic assembly of cullin-RING ubiquitin ligases
Abstract
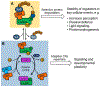
Plants appear quiet: quietly, they break the ground, expand leaves, search for resources, alert each other to invaders, and heal their own wounds. In contrast to the stationary appearance, the inside world of a plant is full of movements: cells divide to increase the body mass and form new organs; signaling molecules migrate among cells and tissues to drive transcriptional cascades and developmental programs; macromolecules, such as RNAs and proteins, collaborate with different partners to maintain optimal organismal function under changing cellular and environmental conditions. All these activities require a dynamic yet appropriately controlled molecular network in plant cells. In this short review, we used the regulation of cullin-RING ubiquitin ligases (CRLs) as an example to discuss how dynamic biochemical processes contribute to plant development. CRLs comprise a large family of modular multi-unit enzymes that determine the activity and stability of diverse regulatory proteins playing crucial roles in plant growth and development. The mechanism governing the dynamic assembly of CRLs is essential for CRL activity and biological function, and it may provide insights and implications for the regulation of other dynamic multi-unit complexes involved in fundamental processes such as transcription, translation, and protein sorting in plants.
Keywords: Adaptive responses; Developmental plasticity; E3 ligases; Molecular dynamics; Ubiquitination.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
Cullin-RING E3 Ubiquitin Ligases: Bridges to Destruction.Subcell Biochem. 2017;83:323-347. doi: 10.1007/978-3-319-46503-6_12. Subcell Biochem. 2017. PMID: 28271482 Free PMC article. Review.
-
Assembly and Regulation of CRL Ubiquitin Ligases.Adv Exp Med Biol. 2020;1217:33-46. doi: 10.1007/978-981-15-1025-0_3. Adv Exp Med Biol. 2020. PMID: 31898220 Review.
-
Cullin 5-RING E3 ubiquitin ligases, new therapeutic targets?Biochimie. 2016 Mar;122:339-47. doi: 10.1016/j.biochi.2015.08.003. Epub 2015 Aug 4. Biochimie. 2016. PMID: 26253693 Review.
-
Structural regulation of cullin-RING ubiquitin ligase complexes.Curr Opin Struct Biol. 2011 Apr;21(2):257-64. doi: 10.1016/j.sbi.2011.01.003. Epub 2011 Feb 1. Curr Opin Struct Biol. 2011. PMID: 21288713 Free PMC article. Review.
-
Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases.Antioxid Redox Signal. 2010 Dec 1;13(11):1699-712. doi: 10.1089/ars.2010.3211. Epub 2010 Aug 14. Antioxid Redox Signal. 2010. PMID: 20486766 Free PMC article. Review.
Cited by
-
Molecular mechanisms of CAND2 in regulating SCF ubiquitin ligases.Nat Commun. 2025 Feb 26;16(1):1998. doi: 10.1038/s41467-025-57065-5. Nat Commun. 2025. PMID: 40011427 Free PMC article.
-
CAND1 inhibits Cullin-2-RING ubiquitin ligases for enhanced substrate specificity.Nat Struct Mol Biol. 2024 Feb;31(2):323-335. doi: 10.1038/s41594-023-01167-5. Epub 2024 Jan 4. Nat Struct Mol Biol. 2024. PMID: 38177676 Free PMC article.
-
Transcriptomic Insight into the Pollen Tube Growth of Olea europaea L. subsp. europaea Reveals Reprogramming and Pollen-Specific Genes Including New Transcription Factors.Plants (Basel). 2023 Aug 8;12(16):2894. doi: 10.3390/plants12162894. Plants (Basel). 2023. PMID: 37631106 Free PMC article.
-
The lowdown on breakdown: Open questions in plant proteolysis.Plant Cell. 2024 Sep 3;36(9):2931-2975. doi: 10.1093/plcell/koae193. Plant Cell. 2024. PMID: 38980154 Free PMC article. Review.
-
The Role of Plant Latex in Virus Biology.Viruses. 2023 Dec 27;16(1):47. doi: 10.3390/v16010047. Viruses. 2023. PMID: 38257746 Free PMC article. Review.
References
-
- ABD-HAMID NA, AHMAD-FAUZI MI, ZAINAL Z & ISMAIL I 2020. Diverse and dynamic roles of F-box proteins in plant biology. Planta, 251, 68. - PubMed
-
- ALONSO-PERAL MM, CANDELA H, DEL POZO JC, MARTINEZ-LABORDA A, PONCE MR & MICOL JL 2006. The HVE/CAND1 gene is required for the early patterning of leaf venation in Arabidopsis. Development, 133, 3755–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

