HLA-E*01:01 + HLA-E*01:01 genotype confers less susceptibility to COVID-19, while HLA-E*01:03 + HLA-E*01:03 genotype is associated with more severe disease
- PMID: 36822912
- PMCID: PMC9922572
- DOI: 10.1016/j.humimm.2023.02.002
HLA-E*01:01 + HLA-E*01:01 genotype confers less susceptibility to COVID-19, while HLA-E*01:03 + HLA-E*01:03 genotype is associated with more severe disease
Abstract
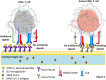
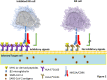
Background: HLA-E interaction with inhibitory receptor, NKG2A attenuates NK-mediated cytotoxicity. NKG2A overexpression by SARS-CoV-2 exhausts NK cells function, whereas virus-induced down-regulation of MHC-Ia reduces its derived-leader sequence peptide levels required for proper binding of HLA-E to NKG2A. This leads HLA-E to become more complex with viral antigens and delivers them to CD8+ T cells, which facilitates cytolysis of infected cells. Now, the fact that alleles of HLA-E have different levels of expression and affinity for MHC Ia-derived peptide raises the question of whether HLA-E polymorphisms affect susceptibility to COVID-19 or its severity.
Methods: 104 COVID-19 convalescent plasma donors with/without history of hospitalization and 18 blood donors with asymptomatic COVID-19, all were positive for anti-SARS-CoV-2 IgG antibody as well as a group of healthy control including 68 blood donors with negative antibody were subjected to HLA-E genotyping. As a privilege, individuals hadn't been vaccinated against COVID-19 and therefore naturally exposed to the SARS-CoV-2.
Results: The absence of HLA-E*01:03 allele significantly decreases the odds of susceptibility to SARS-CoV-2 infection [p = 0.044; OR (95 %CI) = 0.530 (0.286 - 0.983)], suggesting that HLA-E*01:01 + HLA-E*01:01 genotype favors more protection against SARS-CoV-2 infection. HLA-E*01:03 + HLA-E*01:03 genotype was also significantly associated with more severe COVID-19 [p = 0.020; 2.606 (1.163 - 5.844) CONCLUSION: Here, our observation about lower susceptibility of HLA-E*01:01 + HLA-E*01:01 genotype to COVID-19 could be clinical evidence in support of some previous studies suggesting that the lower affinity of HLA-E*01:01 to peptides derived from the leader sequence of MHC class Ia may instead shift its binding to virus-derived peptides, which then facilitates target recognition by restricted conventional CD8+ T cells and leads to efficient cytolysis. On the other hand, according to other studies, less reactivity of HLA-E*01:01 with NKG2A abrogates NK cells or T cells inhibition, which may also lead to a greater cytotoxicity against SARS-CoV-2 infected cells compared to HLA-E*01:03. Taken together given HLA-E polymorphisms, the data presented here may be useful in identifying more vulnerable individuals to COVID-19 for better care and management. Especially since along with other risk factors in patients, having HLA-E*01:03 + HLA-E*01:03 genotype may also be associated with the possibility of severe cases of the disease.
Keywords: CD8(+) T cell; Coronavirus disease 2019 (COVID-19); HLA-E; NK cell; NKG2A; Polymorphism; SARS-CoV-2; Susceptibility.
Copyright © 2023 American Society for Histocompatibility and Immunogenetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
SARS-CoV-2 Nsp13 encodes for an HLA-E-stabilizing peptide that abrogates inhibition of NKG2A-expressing NK cells.Cell Rep. 2022 Mar 8;38(10):110503. doi: 10.1016/j.celrep.2022.110503. Epub 2022 Feb 21. Cell Rep. 2022. PMID: 35235832 Free PMC article.
-
The Impact of Killer Cell Immunoglobulin-Like Receptors and Human Leukocyte Antigen-E, Human Leukocyte Antigen-G Polymorphisms on Innate Immunity and COVID-19 Severity.J Immunol Res. 2025 May 12;2025:6691437. doi: 10.1155/jimr/6691437. eCollection 2025. J Immunol Res. 2025. PMID: 40391199 Free PMC article.
-
HLA-E-restricted SARS-CoV-2-specific T cells from convalescent COVID-19 patients suppress virus replication despite HLA class Ia down-regulation.Sci Immunol. 2023 Jun 30;8(84):eabl8881. doi: 10.1126/sciimmunol.abl8881. Epub 2023 Jun 30. Sci Immunol. 2023. PMID: 37390223
-
HLA Variation and SARS-CoV-2 Specific Antibody Response.Viruses. 2023 Mar 31;15(4):906. doi: 10.3390/v15040906. Viruses. 2023. PMID: 37112884 Free PMC article. Review.
-
Enhancing Natural Killer and CD8+ T Cell-Mediated Anticancer Cytotoxicity and Proliferation of CD8+ T Cells with HLA-E Monospecific Monoclonal Antibodies.Monoclon Antib Immunodiagn Immunother. 2019 Apr;38(2):38-59. doi: 10.1089/mab.2018.0043. Monoclon Antib Immunodiagn Immunother. 2019. PMID: 31009335 Free PMC article. Review.
Cited by
-
Transcriptomic profiling of severe and critical COVID-19 patients reveals alterations in expression, splicing and polyadenylation.Sci Rep. 2025 Apr 18;15(1):13469. doi: 10.1038/s41598-025-95905-y. Sci Rep. 2025. PMID: 40251257 Free PMC article.
-
Non-Classical HLA Class 1b and Hepatocellular Carcinoma.Biomedicines. 2023 Jun 9;11(6):1672. doi: 10.3390/biomedicines11061672. Biomedicines. 2023. PMID: 37371767 Free PMC article. Review.
-
Hantaan virus-derived peptides that stabilize HLA-E could abrogate inhibition of CD56dimNKG2A+ NK cells.PLoS Pathog. 2025 Jul 18;21(7):e1012717. doi: 10.1371/journal.ppat.1012717. eCollection 2025 Jul. PLoS Pathog. 2025. PMID: 40680069 Free PMC article.
-
Exercise: a non-drug strategy of NK cell activation.Braz J Med Biol Res. 2024 Nov 25;57:e14144. doi: 10.1590/1414-431X2024e14144. eCollection 2024. Braz J Med Biol Res. 2024. PMID: 39607207 Free PMC article. Review.
-
Associations Between Clinical Manifestations of SARS-CoV-2 Infection and HLA Alleles in a Caucasian Population: A Molecular HLA Typing Study.J Clin Med. 2024 Dec 17;13(24):7695. doi: 10.3390/jcm13247695. J Clin Med. 2024. PMID: 39768617 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

