Vorasidenib and ivosidenib in IDH1-mutant low-grade glioma: a randomized, perioperative phase 1 trial
- PMID: 36823302
- PMCID: PMC10313524
- DOI: 10.1038/s41591-022-02141-2
Vorasidenib and ivosidenib in IDH1-mutant low-grade glioma: a randomized, perioperative phase 1 trial
Erratum in
-
Author Correction: Vorasidenib and ivosidenib in IDH1-mutant low-grade glioma: a randomized, perioperative phase 1 trial.Nat Med. 2024 Jan;30(1):302. doi: 10.1038/s41591-023-02473-7. Nat Med. 2024. PMID: 37400643 Free PMC article. No abstract available.
Abstract
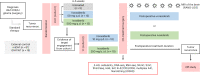
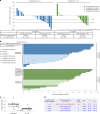
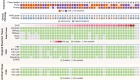
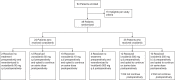
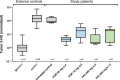
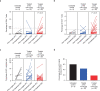
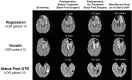
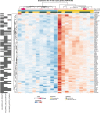
Vorasidenib and ivosidenib inhibit mutant forms of isocitrate dehydrogenase (mIDH) and have shown preliminary clinical activity against mIDH glioma. We evaluated both agents in a perioperative phase 1 trial to explore the mechanism of action in recurrent low-grade glioma (IGG) and select a molecule for phase 3 testing. Primary end-point was concentration of D-2-hydroxyglutarate (2-HG), the metabolic product of mIDH enzymes, measured in tumor tissue from 49 patients with mIDH1-R132H nonenhancing gliomas following randomized treatment with vorasidenib (50 mg or 10 mg once daily, q.d.), ivosidenib (500 mg q.d. or 250 mg twice daily) or no treatment before surgery. Tumor 2-HG concentrations were reduced by 92.6% (95% credible interval (CrI), 76.1-97.6) and 91.1% (95% CrI, 72.0-97.0) in patients treated with vorasidenib 50 mg q.d. and ivosidenib 500 mg q.d., respectively. Both agents were well tolerated and follow-up is ongoing. In exploratory analyses, 2-HG reduction was associated with increased DNA 5-hydroxymethylcytosine, reversal of 'proneural' and 'stemness' gene expression signatures, decreased tumor cell proliferation and immune cell activation. Vorasidenib, which showed brain penetrance and more consistent 2-HG suppression than ivosidenib, was advanced to phase 3 testing in patients with mIDH LGGs. Funded by Agios Pharmaceuticals, Inc. and Servier Pharmaceuticals LLC; ClinicalTrials.gov number NCT03343197.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
The clinical trial was supported by Agios Pharmaceuticals, Inc. Servier Pharmaceuticals LLC completed the acquisition of Agios’ oncology business. I.K.M. reports serving as a consultant for Agios Pharmaceuticals, Black Diamond Therapeutics, Debiopharm Group, Puma Biotechnology, Servier Pharmaceuticals LLC, Voyager Therapeutics, DC Europa Ltd, Kazia Therapeutics, Novartis, Cardinal Health, Roche, Vigeo Therapeutics and Samus Therapeutics. M.L. reports employment with and stock ownership at Agios Pharmaceuticals Inc. and employment with Servier Pharmaceuticals LLC during the conduct of the study. P.Y.W. reports research support from AstraZeneca/MedImmune, Beigene, Celgene, Chimerix, Eli Lily, Genentech/Roche, Kazia, MediciNova, Merck, Novartis, Nuvation Bio, Servier Pharmaceuticals LLC, Vascular Biogenics and VBI Vaccines; and serving on an advisory board for AstraZeneca, Bayer, Black Diamond, Boehringer Ingelheim, Boston Pharmaceuticals, Celularity, Chimerix, Day One Bio, Genenta, GlaxoSmithKline, Karyopharm, Merck, Mundipharma, Novartis, Novocure, Nuvation Bio, Prelude Therapeutics, Sapience, Servier Pharmaceuticals LLC, Sagimet, Vascular Biogenics and VBI Vaccines. J.W.T. reports involvement as a principal investigator and involved in data collection and analysis, and manuscript preparation for clinical trials with BMS, Navio and AbbVie within the last five years. K.B.P. reports research support from Servier Pharmaceuticals LLC. B.M.E. reports serving as a consultant for MedQIA and Servier Pharmaceuticals LLC. K.L. reports employment with and stock ownership at Agios Pharmaceuticals Inc. during the conduct of the study. A.T. reports employment with and stock ownership at Agios Pharmaceuticals Inc. and employment with Servier Pharmaceuticals LLC. S. Choe reports employment with and stock ownership at Agios Pharmaceuticals Inc. and employment with Servier Pharmaceuticals LLC. Y.T. reports employment with and stock ownership at Agios Pharmaceuticals Inc. and employment with Servier Pharmaceuticals LLC during the conduct of the study. S.S. reports employment with and stock ownership at Agios Pharmaceuticals Inc. and employment with Servier Pharmaceuticals LLC. S.S.P. reports employment with and stock ownership at Agios Pharmaceuticals Inc. and employment with Servier Pharmaceuticals LLC. I.H. reports employment with and stock ownership at Agios Pharmaceuticals Inc. and employment with Servier Pharmaceuticals LLC. L.S. reports employment with and stock ownership at Agios Pharmaceuticals Inc. and employment with Servier Pharmaceuticals LLC. J.L.C. reports serving as a consultant for Agios Pharmaceuticals Inc. and Servier Pharmaceuticals LLC. T.F.C. reports being a cofounder, major stock holder, consultant and board member of Katmai Pharmaceuticals, and membership of the board, and paid consultancy for the 501c3 Global Coalition for Adaptive Research; holding stock in Chimerix and receiving milestone payments and possible future royalties; membership of the scientific advisory boards for Break Through Cancer and Cure Brain Cancer Foundation; providing paid consulting services to Sagimet, Clinical Care Options, Ideology Health, Servier, Jubilant, Immvira, Gan & Lee, BrainStorm, Katmai, Sapience, Inovio, Vigeo Therapeutics, DNATrix, Tyme, SDP, Novartis, Roche, Kintara, Bayer, Merck, Boehringer Ingelheim, VBL, Amgen, Kiyatec, Odonate Therapeutics QED, Medefield, Pascal Biosciences, Bayer, Tocagen, Karyopharm, GW Pharma, Abbvie, VBI, Deciphera, VBL, Agios, Genocea, Celgene, Puma, Lilly, BMS, Cortice, Wellcome Trust, Novocure, Novogen, Boston Biomedical, Sunovion, Human Longevity, Insys, ProNai, Pfizer, Notable labs, Medqia Trizel, Medscape; contracts with UCLA for the Brain Tumor Program with Oncovir, Merck, Oncoceutics, Novartis, Amgen, Abbvie, DNAtrix, Beigene, BMS, AstraZeneca, Kazia, Agios, Boston Biomedical, Deciphera, Tocagen, Orbus, AstraZeneca, Karyopharm; and the Regents of the University of California (T.F.C.’s employer) have licensed intellectual property co-invented by T.F.C. to Katmai Pharmaceuticals. No disclosures were reported by the other authors.
Figures





References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

