Network expansion of genetic associations defines a pleiotropy map of human cell biology
- PMID: 36823319
- PMCID: PMC10011132
- DOI: 10.1038/s41588-023-01327-9
Network expansion of genetic associations defines a pleiotropy map of human cell biology
Abstract
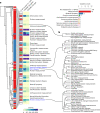
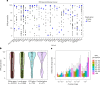
Interacting proteins tend to have similar functions, influencing the same organismal traits. Interaction networks can be used to expand the list of candidate trait-associated genes from genome-wide association studies. Here, we performed network-based expansion of trait-associated genes for 1,002 human traits showing that this recovers known disease genes or drug targets. The similarity of network expansion scores identifies groups of traits likely to share an underlying genetic and biological process. We identified 73 pleiotropic gene modules linked to multiple traits, enriched in genes involved in processes such as protein ubiquitination and RNA processing. In contrast to gene deletion studies, pleiotropy as defined here captures specifically multicellular-related processes. We show examples of modules linked to human diseases enriched in genes with known pathogenic variants that can be used to map targets of approved drugs for repurposing. Finally, we illustrate the use of network expansion scores to study genes at inflammatory bowel disease genome-wide association study loci, and implicate inflammatory bowel disease-relevant genes with strong functional and genetic support.
© 2023. The Author(s).
Conflict of interest statement
C.A.A. has received consultancy fees from Genomics PLC and BridgeBio Inc. G.B. is an employee of GSK. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

