Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4
- PMID: 36823356
- PMCID: PMC10635831
- DOI: 10.1038/s41587-023-01688-w
Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4
Abstract
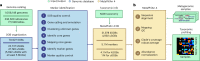
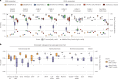
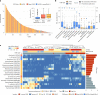
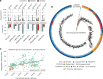
Metagenomic assembly enables new organism discovery from microbial communities, but it can only capture few abundant organisms from most metagenomes. Here we present MetaPhlAn 4, which integrates information from metagenome assemblies and microbial isolate genomes for more comprehensive metagenomic taxonomic profiling. From a curated collection of 1.01 M prokaryotic reference and metagenome-assembled genomes, we define unique marker genes for 26,970 species-level genome bins, 4,992 of them taxonomically unidentified at the species level. MetaPhlAn 4 explains ~20% more reads in most international human gut microbiomes and >40% in less-characterized environments such as the rumen microbiome and proves more accurate than available alternatives on synthetic evaluations while also reliably quantifying organisms with no cultured isolates. Application of the method to >24,500 metagenomes highlights previously undetected species to be strong biomarkers for host conditions and lifestyles in human and mouse microbiomes and shows that even previously uncharacterized species can be genetically profiled at the resolution of single microbial strains.
© 2023. The Author(s).
Conflict of interest statement
S.E.B., T.D.S., F.A. and N.S. are consultants to Zoe Global. F.G, R.D. and J.W. are employees of Zoe Global. The other authors declare no competing interests.
Figures


References
-
- Lu J, Breitwieser FP, Thielen P, Salzberg SL. Bracken: estimating species abundance in metagenomics data. PeerJ Comput. Sci. 2017;3:e104. doi: 10.7717/peerj-cs.104. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

