Proteogenomic links to human metabolic diseases
- PMID: 36823471
- PMCID: PMC7614946
- DOI: 10.1038/s42255-023-00753-7
Proteogenomic links to human metabolic diseases
Erratum in
-
Author Correction: Proteogenomic links to human metabolic diseases.Nat Metab. 2023 Apr;5(4):710. doi: 10.1038/s42255-023-00785-z. Nat Metab. 2023. PMID: 36935425 No abstract available.
Abstract
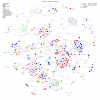
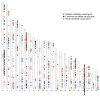
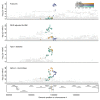
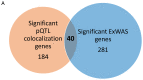
Studying the plasma proteome as the intermediate layer between the genome and the phenome has the potential to identify new disease processes. Here, we conducted a cis-focused proteogenomic analysis of 2,923 plasma proteins measured in 1,180 individuals using antibody-based assays. We (1) identify 256 unreported protein quantitative trait loci (pQTL); (2) demonstrate shared genetic regulation of 224 cis-pQTLs with 575 specific health outcomes, revealing examples for notable metabolic diseases (such as gastrin-releasing peptide as a potential therapeutic target for type 2 diabetes); (3) improve causal gene assignment at 40% (n = 192) of overlapping risk loci; and (4) observe convergence of phenotypic consequences of cis-pQTLs and rare loss-of-function gene burden for 12 proteins, such as TIMD4 for lipoprotein metabolism. Our findings demonstrate the value of integrating complementary proteomic technologies with genomics even at moderate scale to identify new mediators of metabolic diseases with the potential for therapeutic interventions.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
E.W. is now an employee of AstraZeneca. The remaining authors declare no competing interests.
Figures






Comment in
-
Proteogenomic mapping sets stage for precision medicine.Nat Metab. 2023 Mar;5(3):366-367. doi: 10.1038/s42255-023-00759-1. Nat Metab. 2023. PMID: 36823472 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

