Circ_0068631 sponges miR-139-5p to promote the growth and metastasis of cutaneous squamous cell carcinoma by upregulating HOXB7
- PMID: 36823512
- PMCID: PMC10155854
- DOI: 10.1111/srt.13248
Circ_0068631 sponges miR-139-5p to promote the growth and metastasis of cutaneous squamous cell carcinoma by upregulating HOXB7
Abstract
Background: Circular RNAs (circRNAs) are often dysregulated in cancers and closely related to cancer progression, including cutaneous squamous cell carcinoma (CSCC). However, the role and mechanism of circ_0068631 in CSCC progression have not been reported.
Methods: The expression of circ_0068631, microRNA-139-5p (miR-139-5p), and homeobox B7 (HOXB7) was measured by real-time quantitative polymerase chain reaction (RT-qPCR). Cell counting kit-8 (CCK-8) assay, 5-ethynyl-2'-deoxyuridine (EdU) assay, and colony formation assay were used to measure cell proliferation. Cell apoptosis was assessed by flow cytometry. Cell migration was detected by transwell assay. The interaction between miR-139-5p and circ_0068631 or HOXB7 was confirmed by dual-luciferase reporter assay. A xenograft tumor model was established to confirm the function of circ_0068631 in vivo.
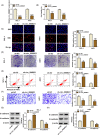
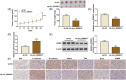
Results: Circ_0068631 was upregulated in CSCC tissues and cells, and its silencing could inhibit CSCC cell proliferation and metastasis while promoting apoptosis in vitro, as well as restrain CSCC tumor growth in vivo. Circ_0068631 acted as a sponge of miR-139-5p, and miR-139-5p inhibition reversed the repressive effect of circ_0068631 knockdown on CSCC cell progression. Furthermore, HOXB7 was a target of miR-139-5p, and miR-139-5p inhibited the malignant behaviors by downregulating HOXB7 expression in CSCC cells. Further, circ_0068631 sponged miR-139-5p to regulate HOXB7 expression.
Conclusion: Circ_0068631 functioned as a novel oncogene in CSCC progression by regulating miR-139-5p/HOXB7 axis, suggesting that circ_0068631 may be a potential target for CSCC treatment.
Highlights: Circ_0068631 was overexpressed in CSCC tissues and cells. Circ_0068631 downregulation suppressed CSCC progression via miR-139-5p. Circ_0068631 regulated HOXB7 via sponging miR-139-5p.
Keywords: HOXB7; circ_0068631; cutaneous squamous cell carcinoma; miR-139-5p.
© 2022 The Authors. Skin Research and Technology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Circular RNA circ-LARP1B contributes to cutaneous squamous cell carcinoma progression by targeting microRNA-515-5p/TPX2 microtubule nucleation factor axis.Bioengineered. 2022 Jan;13(1):1209-1223. doi: 10.1080/21655979.2021.2019172. Bioengineered. 2022. PMID: 34982022 Free PMC article.
-
Hsa_circ_0005085 may suppress cutaneous squamous cell carcinoma growth and metastasis through targeting the miR-186-5p/LAMC1 axis.Skin Res Technol. 2023 Jun;29(6):e13321. doi: 10.1111/srt.13321. Skin Res Technol. 2023. Retraction in: Skin Res Technol. 2025 Feb-May;31(2-5):e70181. doi: 10.1111/srt.70181. PMID: 37357644 Free PMC article. Retracted.
-
CircRNA circ_0067772 aggravates the malignant progression of cutaneous squamous cell carcinoma by regulating miR-1238-3p/FOXG1 axis.Genes Genomics. 2021 May;43(5):491-501. doi: 10.1007/s13258-021-01074-3. Epub 2021 Mar 11. Genes Genomics. 2021. PMID: 33709381
-
Circ_0001402 knockdown suppresses the chemoresistance and development of DDP-resistant cutaneous squamous cell carcinoma cells by functioning as a ceRNA for miR-625-5p.Exp Dermatol. 2023 Apr;32(4):529-541. doi: 10.1111/exd.14745. Epub 2023 Jan 31. Exp Dermatol. 2023. PMID: 36635223
-
Circ-CSPP1 knockdown suppresses hepatocellular carcinoma progression through miR-493-5p releasing-mediated HMGB1 downregulation.Cell Signal. 2021 Oct;86:110065. doi: 10.1016/j.cellsig.2021.110065. Epub 2021 Jun 26. Cell Signal. 2021. PMID: 34182091 Review.
Cited by
-
Circular RNA TFRC/SCD1 mRNA interaction regulates ferroptosis and metastasis in gastric cancer.Cell Death Dis. 2025 Jun 5;16(1):436. doi: 10.1038/s41419-025-07759-x. Cell Death Dis. 2025. PMID: 40473597 Free PMC article.
References
-
- Que SKT, Zwald FO, Schmults CD. Cutaneous squamous cell carcinoma: incidence, risk factors, diagnosis, and staging. J Am Acad Dermatol. 2018;78(2):237‐247. - PubMed
-
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333‐338. - PubMed
-
- Yao T, Chen Q, Fu L, Guo J. Circular RNAs: biogenesis, properties, roles, and their relationships with liver diseases. Hepatol Res. 2017;47(6):497‐504. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

