Transition Substitution of Desired Bases in Human Pluripotent Stem Cells with Base Editors: A Step-by-Step Guide
- PMID: 36823978
- PMCID: PMC10226858
- DOI: 10.15283/ijsc22171
Transition Substitution of Desired Bases in Human Pluripotent Stem Cells with Base Editors: A Step-by-Step Guide
Abstract
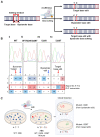
The recent advances in human pluripotent stem cells (hPSCs) enable to precisely edit the desired bases in hPSCs to be used for the establishment of isogenic disease models and autologous ex vivo cell therapy. The knock-in approach based on the homologous directed repair with Cas9 endonuclease, causing DNA double-strand breaks (DSBs), produces not only insertion and deletion (indel) mutations but also deleterious large deletions. On the contrary, due to the lack of Cas9 endonuclease activity, base editors (BEs) such as adenine base editor (ABE) and cytosine base editor (CBE) allow precise base substitution by conjugated deaminase activity, free from DSB formation. Despite the limitation of BEs in transition substitution, precise base editing by BEs with no massive off-targets is suggested to be a prospective alternative in hPSCs for clinical applications. Considering the unique cellular characteristics of hPSCs, a few points should be considered. Herein, we describe an updated and optimized protocol for base editing in hPSCs. We also describe an improved methodology for CBE-based C to T substitutions, which are generally lower than A to G substitutions in hPSCs.
Keywords: Base editors; Cas9; Genome editing; Human pluripotent stem cells; Transition substitution.
Conflict of interest statement
The authors have no conflicting financial interest.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials

