Efficient and stable metabarcoding sequencing data using a DNBSEQ-G400 sequencer validated by comprehensive community analyses
- PMID: 36824325
- PMCID: PMC9632034
- DOI: 10.46471/gigabyte.16
Efficient and stable metabarcoding sequencing data using a DNBSEQ-G400 sequencer validated by comprehensive community analyses
Abstract
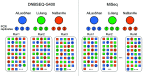
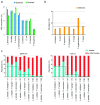
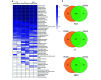
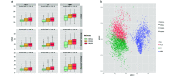
Metabarcoding is a widely used method for fast characterization of microbial communities in complex environmental samples. However, the selction of sequencing platform can have a noticeable effect on the estimated community composition. Here, we evaluated the metabarcoding performance of a DNBSEQ-G400 sequencer developed by MGI Tech using 16S and internal transcribed spacer (ITS) markers to investigate bacterial and fungal mock communities, as well as the ITS2 marker to investigate the fungal community of 1144 soil samples, with additional technical replicates. We show that highly accurate sequencing of bacterial and fungal communities is achievable using DNBSEQ-G400. Measures of diversity and correlation from soil metabarcoding showed that the results correlated highly with those of different machines of the same model, as well as between different sequencing modes (single-end 400 bp and paired-end 200 bp). Moderate, but significant differences were observed between results produced with different sequencing platforms (DNBSEQ-G400 and MiSeq); however, the highest differences can be caused by selecting different primer pairs for PCR amplification of taxonomic markers. These differences suggested that care is needed while jointly analyzing metabarcoding data from differenet experiments. This study demonstrated the high performance and accuracy of DNBSEQ-G400 for short-read metabarcoding of microbial communities. Our study also produced datasets to allow further investigation of microbial diversity.
© The Author(s) 2021.
Figures


Similar articles
-
Highly comparable metabarcoding results from MGI-Tech and Illumina sequencing platforms.PeerJ. 2021 Sep 30;9:e12254. doi: 10.7717/peerj.12254. eCollection 2021. PeerJ. 2021. PMID: 34703674 Free PMC article.
-
Sequencing introduced false positive rare taxa lead to biased microbial community diversity, assembly, and interaction interpretation in amplicon studies.Environ Microbiome. 2022 Aug 17;17(1):43. doi: 10.1186/s40793-022-00436-y. Environ Microbiome. 2022. PMID: 35978448 Free PMC article.
-
Benchmarking of ATAC Sequencing Data From BGI's Low-Cost DNBSEQ-G400 Instrument for Identification of Open and Occupied Chromatin Regions.Front Mol Biosci. 2022 Jul 7;9:900323. doi: 10.3389/fmolb.2022.900323. eCollection 2022. Front Mol Biosci. 2022. PMID: 35874611 Free PMC article.
-
Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies.PLoS One. 2014 Jun 16;9(6):e97629. doi: 10.1371/journal.pone.0097629. eCollection 2014. PLoS One. 2014. PMID: 24933453 Free PMC article.
-
DNA Metabarcoding for the Characterization of Terrestrial Microbiota-Pitfalls and Solutions.Microorganisms. 2021 Feb 12;9(2):361. doi: 10.3390/microorganisms9020361. Microorganisms. 2021. PMID: 33673098 Free PMC article. Review.
Cited by
-
High-resolution single-molecule long-fragment rRNA gene amplicon sequencing of bacterial and eukaryotic microbial communities.Cell Rep Methods. 2023 Mar 27;3(3):100437. doi: 10.1016/j.crmeth.2023.100437. eCollection 2023 Mar 27. Cell Rep Methods. 2023. PMID: 37056375 Free PMC article.
-
Highly comparable metabarcoding results from MGI-Tech and Illumina sequencing platforms.PeerJ. 2021 Sep 30;9:e12254. doi: 10.7717/peerj.12254. eCollection 2021. PeerJ. 2021. PMID: 34703674 Free PMC article.
-
Unique Nucleotide Polymorphism of African Swine Fever Virus Circulating in East Asia and Central Russia.Viruses. 2024 Dec 11;16(12):1907. doi: 10.3390/v16121907. Viruses. 2024. PMID: 39772214 Free PMC article.
-
Sequencing introduced false positive rare taxa lead to biased microbial community diversity, assembly, and interaction interpretation in amplicon studies.Environ Microbiome. 2022 Aug 17;17(1):43. doi: 10.1186/s40793-022-00436-y. Environ Microbiome. 2022. PMID: 35978448 Free PMC article.
-
Multi-Omics Analysis Revealed the Accumulation of Flavonoids and Shift of Fungal Community Structure Caused by Tea Grafting (Camellia sinensis L.).Plants (Basel). 2025 Apr 10;14(8):1176. doi: 10.3390/plants14081176. Plants (Basel). 2025. PMID: 40284064 Free PMC article.
References
LinkOut - more resources
Full Text Sources
