Improvements in the sequencing and assembly of plant genomes
- PMID: 36824328
- PMCID: PMC9631998
- DOI: 10.46471/gigabyte.24
Improvements in the sequencing and assembly of plant genomes
Abstract
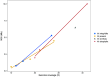
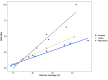
Advances in DNA sequencing have made it easier to sequence and assemble plant genomes. Here, we extend an earlier study, and compare recent methods for long read sequencing and assembly. Updated Oxford Nanopore Technology software improved assemblies. Using more accurate sequences produced by repeated sequencing of the same molecule (Pacific Biosciences HiFi) resulted in less fragmented assembly of sequencing reads. Using data for increased genome coverage resulted in longer contigs, but reduced total assembly length and improved genome completeness. The original model species, Macadamia jansenii, was also compared with three other Macadamia species, as well as avocado (Persea americana) and jojoba (Simmondsia chinensis). In these angiosperms, increasing sequence data volumes caused a linear increase in contig size, decreased assembly length and further improved already high completeness. Differences in genome size and sequence complexity influenced the success of assembly. Advances in long read sequencing technology continue to improve plant genome sequencing and assembly. However, results were improved by greater genome coverage, with the amount needed to achieve a particular level of assembly being species dependent.
© The Author(s) 2021.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Comment on
-
Comparison of long-read methods for sequencing and assembly of a plant genome.Gigascience. 2020 Dec 21;9(12):giaa146. doi: 10.1093/gigascience/giaa146. Gigascience. 2020. PMID: 33347571 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
