This is a preprint.
Accelerated evolution of SARS-CoV-2 in free-ranging white-tailed deer
- PMID: 36824718
- PMCID: PMC9949239
- DOI: 10.21203/rs.3.rs-2574993/v1
Accelerated evolution of SARS-CoV-2 in free-ranging white-tailed deer
Update in
-
Accelerated evolution of SARS-CoV-2 in free-ranging white-tailed deer.Nat Commun. 2023 Aug 28;14(1):5105. doi: 10.1038/s41467-023-40706-y. Nat Commun. 2023. PMID: 37640694 Free PMC article.
Abstract
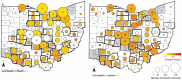
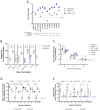
While SARS-CoV-2 has sporadically infected a wide range of animal species worldwide1, the virus has been repeatedly and frequently detected in white-tailed deer in North America2â€"7. The zoonotic origins of this pandemic virus highlight the need to fill the vast gaps in our knowledge of SARS-CoV-2 ecology and evolution in non-human hosts. Here, we detected SARS-CoV-2 was introduced from humans into white-tailed deer more than 30 times in Ohio, USA during November 2021-March 2022. Subsequently, deer-to-deer transmission persisted for 2-8 months, which disseminated across hundreds of kilometers. We discovered that alpha and delta variants evolved in white-tailed deer at three-times the rate observed in humans. Newly developed Bayesian phylogenetic methods quantified how SARS-CoV-2 evolution is not only faster in white-tailed deer but driven by different mutational biases and selection pressures. White-tailed deer are not just short-term recipients of human viral diversity but serve as reservoirs for alpha and other variants to evolve in new directions after going extinct in humans. The long-term effect of this accelerated evolutionary rate remains to be seen as no critical phenotypic changes were observed in our animal model experiments using viruses isolated from white-tailed deer. Still, SARS-CoV-2 viruses have transmitted in white-tailed deer populations for a relatively short duration, and the risk of future changes may have serious consequences for humans and livestock.
Conflict of interest statement
Figures



References
-
- World Organisation for Animal Health. SARS-CoV-2 in Animals - Situation Report 20. https://www.woah.org/app/uploads/2023/01/sars-cov-2-situation-report-20.pdf (2022).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

