This is a preprint.
Direct long read visualization reveals metabolic interplay between two antimalarial drug targets
- PMID: 36824743
- PMCID: PMC9948948
- DOI: 10.1101/2023.02.13.528367
Direct long read visualization reveals metabolic interplay between two antimalarial drug targets
Abstract
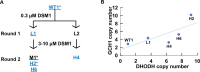
Increases in the copy number of large genomic regions, termed genome amplification, are an important adaptive strategy for malaria parasites. Numerous amplifications across the Plasmodium falciparum genome contribute directly to drug resistance or impact the fitness of this protozoan parasite. During the characterization of parasite lines with amplifications of the dihydroorotate dehydrogenase (DHODH) gene, we detected increased copies of an additional genomic region that encompassed 3 genes (~5 kb) including GTP cyclohydrolase I (GCH1 amplicon). While this gene is reported to increase the fitness of antifolate resistant parasites, GCH1 amplicons had not previously been implicated in any other antimalarial resistance context. Here, we further explored the association between GCH1 and DHODH copy number. Using long read sequencing and single read visualization, we directly observed a higher number of tandem GCH1 amplicons in parasites with increased DHODH copies (up to 9 amplicons) compared to parental parasites (3 amplicons). While all GCH1 amplicons shared a consistent structure, expansions arose in 2-unit steps (from 3 to 5 to 7, etc copies). Adaptive evolution of DHODH and GCH1 loci was further bolstered when we evaluated prior selection experiments; DHODH amplification was only successful in parasite lines with pre-existing GCH1 amplicons. These observations, combined with the direct connection between metabolic pathways that contain these enzymes, lead us to propose that the GCH1 locus is beneficial for the fitness of parasites exposed to DHODH inhibitors. This finding highlights the importance of studying variation within individual parasite genomes as well as biochemical connections of drug targets as novel antimalarials move towards clinical approval.
Figures




References
-
- Casares S, Brumeanu TD, Richie TL. The RTS,S malaria vaccine. Vaccine. 2010. Jul 12;28(31):4880–94. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
