This is a preprint.
Genetic mechanisms of 184 neuro-related proteins in human plasma
- PMID: 36824751
- PMCID: PMC9949195
- DOI: 10.1101/2023.02.10.23285650
Genetic mechanisms of 184 neuro-related proteins in human plasma
Update in
-
The genetic landscape of neuro-related proteins in human plasma.Nat Hum Behav. 2024 Nov;8(11):2222-2234. doi: 10.1038/s41562-024-01963-z. Epub 2024 Aug 29. Nat Hum Behav. 2024. PMID: 39210026
Abstract
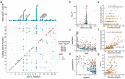
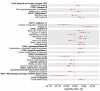
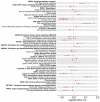
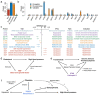
Understanding the genetic basis of neuro-related proteins is essential for dissecting the disease etiology of neuropsychiatric disorders and other complex traits and diseases. Here, the SCALLOP Consortium conducted a genome-wide association meta-analysis of over 12,500 individuals for 184 neuro-reiated proteins in human plasma. The analysis identified 117 cis-regulatory protein quantitative trait loci (cis-pQTL) and 166 trans-pQTL. The mapped pQTL capture on average 50% of each protein's heritability. Mendelian randomization analyses revealed multiple proteins showing potential causal effects on neuro-reiated traits as well as complex diseases such as hypertension, high cholesterol, immune-related disorders, and psychiatric disorders. Integrating with established drug information, we validated 13 combinations of protein targets and diseases or side effects with available drugs, while suggesting hundreds of re-purposing and new therapeutic targets for diseases and comorbidities. This consortium effort provides a large-scale proteogenomic resource for biomedical research.
Conflict of interest statement
Competing interests statement P.R.H.J.T is a salaried employee of BioAge Labs, Inc. The remaining authors declare no competing financial interests. R.E.M has received a speaker fee from Illumina, is an advisor to the Epigenetic Clock Development Foundation, and a scientific consultant for Optima Partners. E.W. is now an employee of AstraZeneca.
Figures

References
-
- WHO. Who | mental disorders (2019). URL https://www.who.int/news-room/fact-sheets/detail/mental-disorders.
-
- Ritchie H. & Roser M. Mental health (2020). URL https://ourworldindata.org/mental-health.
-
- Hossain Μ. M. et al. Epidemiology of mental health problems in covid-19: a review. FlOOOResearch 9 (2020). URL /pmc/articles/PMC7549174//pmc/articles/PMC7549174/report=abstracthttps:/.... - PMC - PubMed
-
- Greenberg N. Mental health of health-care workers in the covid-19 era. Nature Reviews Nephrology 2020 16:8 16, 425–426 (2020). URL https://www.nature.com/articles/s41581-020-0314-5. - PMC - PubMed
-
- Jones E. A., Mitra A. K. & Bhuiyan A. R. Impact of covid-19 on mental health in adolescents: A systematic review. International Journal of Environmental Research and Public Health 2021, Vol. 18, Page 2470 18, 2470 (2021). URL https://www.mdpi.com/1660-4601/18/5/2470/htmhttps://www.mdpi.com/1660-46.... - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
