This is a preprint.
The Pathfinder plasmid toolkit for genetically engineering newly isolated bacteria enables the study of Drosophila -colonizing Orbaceae
- PMID: 36824770
- PMCID: PMC9949093
- DOI: 10.1101/2023.02.15.528778
The Pathfinder plasmid toolkit for genetically engineering newly isolated bacteria enables the study of Drosophila -colonizing Orbaceae
Update in
-
The Pathfinder plasmid toolkit for genetically engineering newly isolated bacteria enables the study of Drosophila-colonizing Orbaceae.ISME Commun. 2023 May 24;3(1):49. doi: 10.1038/s43705-023-00255-3. ISME Commun. 2023. PMID: 37225918 Free PMC article.
Abstract
Toolkits of plasmids and genetic parts streamline the process of assembling DNA constructs and engineering microbes. Many of these kits were designed with specific industrial or laboratory microbes in mind. For researchers interested in non-model microbial systems, it is often unclear which tools and techniques will function in newly isolated strains. To address this challenge, we designed the Pathfinder toolkit for quickly determining the compatibility of a bacterium with different plasmid components. Pathfinder plasmids combine three different broad-host-range origins of replication with multiple antibiotic resistance cassettes and reporters, so that sets of parts can be rapidly screened through multiplex conjugation. We first tested these plasmids in Escherichia coli , a strain of Sodalis praecaptivus that colonizes insects, and a Rosenbergiella isolate from leafhoppers. Then, we used the Pathfinder plasmids to engineer previously unstudied bacteria from the family Orbaceae that were isolated from several fly species. Engineered Orbaceae strains were able to colonize Drosophila melanogaster and could be visualized in fly guts. Orbaceae are common and abundant in the guts of wild-caught flies but have not been included in laboratory studies of how the Drosophila microbiome affects fly health. Thus, this work provides foundational genetic tools for studying new host-associated microbes, including bacteria that are a key constituent of the gut microbiome of a model insect species.
Importance: To fully understand how microbes have evolved to interact with their environments, one must be able to modify their genomes. However, it can be difficult and laborious to discover which genetic tools and approaches work for a new isolate. Bacteria from the recently described Orbaceae family are common in the microbiomes of insects. We developed the Pathfinder plasmid toolkit for testing the compatibility of different genetic parts with newly cultured bacteria. We demonstrate its utility by engineering Orbaceae strains isolated from flies to express fluorescent proteins and characterizing how they colonize the Drosophila melanogaster gut. Orbaceae are widespread in Drosophila in the wild but have not been included in laboratory studies examining how the gut microbiome affects fly nutrition, health, and longevity. Our work establishes a path for genetic studies aimed at understanding and altering interactions between these and other newly isolated bacteria and their hosts.
Figures


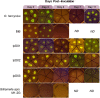
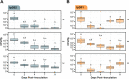

Similar articles
-
The Pathfinder plasmid toolkit for genetically engineering newly isolated bacteria enables the study of Drosophila-colonizing Orbaceae.ISME Commun. 2023 May 24;3(1):49. doi: 10.1038/s43705-023-00255-3. ISME Commun. 2023. PMID: 37225918 Free PMC article.
-
Correction to: The Pathfinder plasmid toolkit for genetically engineering newly isolated bacteria enables the study of Drosophila-colonizing Orbaceae.ISME Commun. 2023 Nov 16;3(1):119. doi: 10.1038/s43705-023-00329-2. ISME Commun. 2023. PMID: 37974005 Free PMC article. No abstract available.
-
Genetic Engineering of Bee Gut Microbiome Bacteria with a Toolkit for Modular Assembly of Broad-Host-Range Plasmids.ACS Synth Biol. 2018 May 18;7(5):1279-1290. doi: 10.1021/acssynbio.7b00399. Epub 2018 Apr 13. ACS Synth Biol. 2018. PMID: 29608282 Free PMC article.
-
Methods of DNA introduction for the engineering of commensal microbes.Eng Microbiol. 2022 Sep 12;2(4):100048. doi: 10.1016/j.engmic.2022.100048. eCollection 2022 Dec. Eng Microbiol. 2022. PMID: 39628703 Free PMC article. Review.
-
Emerging methylation-based approaches in microbiome engineering.Biotechnol Biofuels Bioprod. 2024 Jul 10;17(1):96. doi: 10.1186/s13068-024-02529-x. Biotechnol Biofuels Bioprod. 2024. PMID: 38987811 Free PMC article. Review.
References
-
- Elston KM, Leonard SP, Geng P, Bialik SB, Robinson E, Barrick JE. 2022. Engineering insects from the endosymbiont out. Trends Microbiol 30:79–96. - PubMed
-
- Brophy JAN, Triassi AJ, Adams BL, Renberg RL, Stratis-Cullum DN, Grossman AD, Voigt CA. 2018. Engineered integrative and conjugative elements for efficient and inducible DNA transfer to undomesticated bacteria. Nat Microbiol 3:1043–1053. - PubMed
-
- Durante-Rodríguez G, De Lorenzo V, Martínez-García E. 2014. The standard European vector architecture (SEVA) plasmid toolkit. Methods Mol Biol 1149:469–478. - PubMed
-
- Iverson S V., Haddock TL, Beal J, Densmore DM. 2016. CIDAR MoClo: Improved MoClo assembly standard and new E. coli part library enable rapid combinatorial design for synthetic and traditional biology. ACS Synth Biol 5:99–103. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
