This is a preprint.
The Embryonic Origin of Primordial Germ Cells in the Tardigrade Hypsibius exemplaris
- PMID: 36824831
- PMCID: PMC9948961
- DOI: 10.1101/2023.01.02.522500
The Embryonic Origin of Primordial Germ Cells in the Tardigrade Hypsibius exemplaris
Update in
-
The embryonic origin of primordial germ cells in the tardigrade Hypsibius exemplaris.Dev Biol. 2023 May;497:42-58. doi: 10.1016/j.ydbio.2023.02.008. Epub 2023 Mar 7. Dev Biol. 2023. PMID: 36893882
Abstract
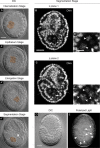
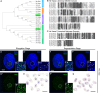
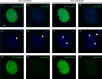
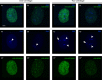
Primordial germ cells (PGCs) give rise to gametes â€" cells necessary for the propagation and fertility of diverse organisms. Current understanding of PGC development is limited to the small number of organisms whose PGCs have been identified and studied. Expanding the field to include little-studied taxa and emerging model organisms is important to understand the full breadth of the evolution of PGC development. In the phylum Tardigrada, no early cell lineages have been identified to date using molecular markers. This includes the PGC lineage. Here, we describe PGC development in the model tardigrade Hypsibius exemplaris . The four earliest-internalizing cells (EICs) exhibit PGC-like behavior and nuclear morphology. The location of the EICs is enriched for mRNAs of conserved PGC markers wiwi1 (water bear piwi 1) and vasa . At early stages, both wiwi1 and vasa mRNAs are detectable uniformly in embryos, which suggests that these mRNAs do not serve as localized determinants for PGC specification. Only later are wiwi1 and vasa enriched in the EICs. Finally, we traced the cells that give rise to the four PGCs. Our results reveal the embryonic origin of the PGCs of H. exemplaris and provide the first molecular characterization of an early cell lineage in the tardigrade phylum. We anticipate that these observations will serve as a basis for characterizing the mechanisms of PGC development in this animal.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
