This is a preprint.
Tumor Niche Network-Defined Subtypes Predict Immunotherapy Response of Esophageal Squamous Cell Cancer
- PMID: 36824935
- PMCID: PMC9949073
- DOI: 10.1101/2023.02.15.528539
Tumor Niche Network-Defined Subtypes Predict Immunotherapy Response of Esophageal Squamous Cell Cancer
Update in
-
Tumor niche network-defined subtypes predict immunotherapy response of esophageal squamous cell cancer.iScience. 2024 Apr 22;27(5):109795. doi: 10.1016/j.isci.2024.109795. eCollection 2024 May 17. iScience. 2024. PMID: 38741711 Free PMC article.
Abstract
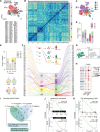
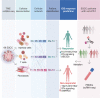
Despite the promising outcomes of immune checkpoint blockade (ICB), resistance to ICB presents a new challenge. Therefore, selecting patients for specific ICB applications is crucial for maximizing therapeutic efficacy. Herein we curated 69 human esophageal squamous cell cancer (ESCC) patients' tumor microenvironment (TME) single-cell transcriptomic datasets to subtype ESCC. Integrative analyses of the cellular network transcriptional signatures of T cells, myeloid cells, and fibroblasts define distinct ESCC subtypes characterized by T cell exhaustion, Interferon (IFN) a/b signaling, TIGIT enrichment, and specific marker genes. Furthermore, this approach classifies ESCC patients into ICB responders and non-responders, as validated by liquid biopsy single-cell transcriptomics. Our study stratifies ESCC patients based on TME transcriptional network, providing novel insights into tumor niche remodeling and predicting ICB responses in ESCC patients.
Keywords: Esophageal squamous cell cancer; cancer immunotherapy; immune checkpoint inhibitors; immunotherapy resistance; single-cell transcriptomics; tumor microenvironment (TME).
Conflict of interest statement
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
Figures






References
-
- Baba Y, Yoshida N, Kinoshita K, Iwatsuki M, Yamashita YI, Chikamoto A, et al. Clinical and Prognostic Features of Patients With Esophageal Cancer and Multiple Primary Cancers: A Retrospective Single-institution Study. Ann Surg. 2018;267(3):478–83. Epub 2017/02/06. doi: 10.1097/SLA.0000000000002118. - DOI - PubMed
-
- Board PATE. Esophageal Cancer Treatment (Adult) (PDQ(R)): Patient Version. PDQ Cancer Information Summaries. Bethesda (MD)2002.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
