Mitochondrial function-associated genes underlie cortical atrophy in prodromal synucleinopathies
- PMID: 36826230
- PMCID: PMC10393413
- DOI: 10.1093/brain/awad044
Mitochondrial function-associated genes underlie cortical atrophy in prodromal synucleinopathies
Abstract
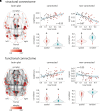
Isolated rapid eye movement sleep behaviour disorder (iRBD) is a sleep disorder characterized by the loss of rapid eye movement sleep muscle atonia and the appearance of abnormal movements and vocalizations during rapid eye movement sleep. It is a strong marker of incipient synucleinopathy such as dementia with Lewy bodies and Parkinson's disease. Patients with iRBD already show brain changes that are reminiscent of manifest synucleinopathies including brain atrophy. However, the mechanisms underlying the development of this atrophy remain poorly understood. In this study, we performed cutting-edge imaging transcriptomics and comprehensive spatial mapping analyses in a multicentric cohort of 171 polysomnography-confirmed iRBD patients [67.7 ± 6.6 (49-87) years; 83% men] and 238 healthy controls [66.6 ± 7.9 (41-88) years; 77% men] with T1-weighted MRI to investigate the gene expression and connectivity patterns associated with changes in cortical thickness and surface area in iRBD. Partial least squares regression was performed to identify the gene expression patterns underlying cortical changes in iRBD. Gene set enrichment analysis and virtual histology were then done to assess the biological processes, cellular components, human disease gene terms, and cell types enriched in these gene expression patterns. We then used structural and functional neighbourhood analyses to assess whether the atrophy patterns in iRBD were constrained by the brain's structural and functional connectome. Moreover, we used comprehensive spatial mapping analyses to assess the specific neurotransmitter systems, functional networks, cytoarchitectonic classes, and cognitive brain systems associated with cortical changes in iRBD. All comparisons were tested against null models that preserved spatial autocorrelation between brain regions and compared to Alzheimer's disease to assess the specificity of findings to synucleinopathies. We found that genes involved in mitochondrial function and macroautophagy were the strongest contributors to the cortical thinning occurring in iRBD. Moreover, we demonstrated that cortical thinning was constrained by the brain's structural and functional connectome and that it mapped onto specific networks involved in motor and planning functions. In contrast with cortical thickness, changes in cortical surface area were related to distinct genes, namely genes involved in the inflammatory response, and to different spatial mapping patterns. The gene expression and connectivity patterns associated with iRBD were all distinct from those observed in Alzheimer's disease. In summary, this study demonstrates that the development of brain atrophy in synucleinopathies is constrained by specific genes and networks.
Keywords: MRI; Parkinson’s disease; REM sleep behaviour disorder; dementia with Lewy bodies; network analysis; transcriptomics.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
None of the authors report any competing interests related to the current work.
Figures



References
-
- Hogl B, Stefani A, Videnovic A. Idiopathic REM sleep behaviour disorder and neurodegeneration—an update. Nat Rev Neurol. 2018;14:40–55. - PubMed
-
- Bourgouin PA, Rahayel S, Gaubert M, et al. Neuroimaging of rapid eye movement sleep behavior disorder. Int Rev Neurobiol. 2019;144:185–210. - PubMed
-
- Campabadal A, Segura B, Junque C, Iranzo A. Structural and functional magnetic resonance imaging in isolated REM sleep behavior disorder: A systematic review of studies using neuroimaging software. Sleep Med Rev. 2021;59:101495. - PubMed
-
- Rahayel S, Postuma RB, Montplaisir J, et al. Abnormal gray matter shape, thickness, and volume in the motor cortico-subcortical loop in idiopathic rapid eye movement sleep behavior disorder: Association with clinical and motor features. Cereb Cortex. 2018;28:658–671. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

