Cardiovascular and Renal Comorbidities Included into Neural Networks Predict the Outcome in COVID-19 Patients Admitted to an Intensive Care Unit: Three-Center, Cross-Validation, Age- and Sex-Matched Study
- PMID: 36826535
- PMCID: PMC9967447
- DOI: 10.3390/jcdd10020039
Cardiovascular and Renal Comorbidities Included into Neural Networks Predict the Outcome in COVID-19 Patients Admitted to an Intensive Care Unit: Three-Center, Cross-Validation, Age- and Sex-Matched Study
Abstract
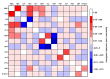
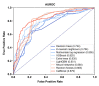
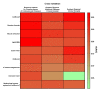
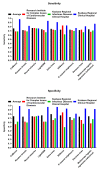
Here, we performed a multicenter, age- and sex-matched study to compare the efficiency of various machine learning algorithms in the prediction of COVID-19 fatal outcomes and to develop sensitive, specific, and robust artificial intelligence tools for the prompt triage of patients with severe COVID-19 in the intensive care unit setting. In a challenge against other established machine learning algorithms (decision trees, random forests, extra trees, neural networks, k-nearest neighbors, and gradient boosting: XGBoost, LightGBM, and CatBoost) and multivariate logistic regression as a reference, neural networks demonstrated the highest sensitivity, sufficient specificity, and excellent robustness. Further, neural networks based on coronary artery disease/chronic heart failure, stage 3-5 chronic kidney disease, blood urea nitrogen, and C-reactive protein as the predictors exceeded 90% sensitivity and 80% specificity, reaching AUROC of 0.866 at primary cross-validation and 0.849 at secondary cross-validation on virtual samples generated by the bootstrapping procedure. These results underscore the impact of cardiovascular and renal comorbidities in the context of thrombotic complications characteristic of severe COVID-19. As aforementioned predictors can be obtained from the case histories or are inexpensive to be measured at admission to the intensive care unit, we suggest this predictor composition is useful for the triage of critically ill COVID-19 patients.
Keywords: C-reactive protein; COVID-19; blood urea nitrogen; chronic kidney disease; coronary artery disease; lymphocyte count; machine learning; neural networks; neutrophil-to-lymphocyte ratio; prognostication.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

References
-
- WHO Coronavirus (COVID-19) Dashboard. [(accessed on 27 November 2022)]. Available online: https://covid19.who.int/
-
- Kamran F., Tang S., Otles E., McEvoy D.S., Saleh S.N., Gong J., Li B.Y., Dutta S., Liu X., Medford R.J., et al. Early identification of patients admitted to hospital for COVID-19 at risk of clinical deterioration: Model development and multisite external validation study. BMJ. 2022;376:e068576. doi: 10.1136/bmj-2021-068576. - DOI - PMC - PubMed
-
- Rasmy L., Nigo M., Kannadath B.S., Xie Z., Mao B., Patel K., Zhou Y., Zhang W., Ross A., Xu H., et al. Recurrent neural network models (CovRNN) for predicting outcomes of patients with COVID-19 on admission to hospital: Model development and validation using electronic health record data. Lancet Digit. Health. 2022;4:e415–e425. doi: 10.1016/S2589-7500(22)00049-8. - DOI - PMC - PubMed
-
- Jiao Z., Choi J.W., Halsey K., Tran T.M.L., Hsieh B., Wang D., Eweje F., Wang R., Chang K., Wu J., et al. Prognostication of patients with COVID-19 using artificial intelligence based on chest x-rays and clinical data: A retrospective study. Lancet Digit. Health. 2021;3:e286–e294. doi: 10.1016/S2589-7500(21)00039-X. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

