Characterization of the Native Disulfide Isomers of the Novel χ-Conotoxin PnID: Implications for Further Increasing Conotoxin Diversity
- PMID: 36827103
- PMCID: PMC9964023
- DOI: 10.3390/md21020061
Characterization of the Native Disulfide Isomers of the Novel χ-Conotoxin PnID: Implications for Further Increasing Conotoxin Diversity
Abstract
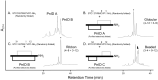
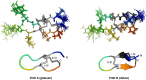
χ-Conotoxins are known for their ability to selectively inhibit norepinephrine transporters, an ability that makes them potential leads for treating various neurological disorders, including neuropathic pain. PnID, a peptide isolated from the venom of Conus pennaceus, shares high sequence homology with previously characterized χ-conotoxins. Whereas previously reported χ-conotoxins seem to only have a single native disulfide bonding pattern, PnID has three native isomers due to the formation of different disulfide bond patterns during its maturation in the venom duct. In this study, the disulfide connectivity and three-dimensional structure of these disulfide isomers were explored using regioselective synthesis, chromatographic coelution, and solution-state nuclear magnetic resonance spectroscopy. Of the native isomers, only the isomer with a ribbon disulfide configuration showed pharmacological activity similar to other χ-conotoxins. This isomer inhibited the rat norepinephrine transporter (IC50 = 10 ± 2 µM) and has the most structural similarity to previously characterized χ-conotoxins. In contrast, the globular isoform of PnID showed more than ten times less activity against this transporter and the beaded isoform did not display any measurable biological activity. This study is the first report of the pharmacological and structural characterization of an χ-conotoxin from a species other than Conus marmoreus and is the first report of the existence of natively-formed conotoxin isomers.
Keywords: conotoxins; isomers; monoamine transporters; peptide; structure; toxins.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the result.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

