Droplet digital PCR-based testing for donor-derived cell-free DNA in transplanted patients as noninvasive marker of allograft health: Methodological aspects
- PMID: 36827438
- PMCID: PMC9955980
- DOI: 10.1371/journal.pone.0282332
Droplet digital PCR-based testing for donor-derived cell-free DNA in transplanted patients as noninvasive marker of allograft health: Methodological aspects
Abstract
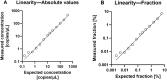
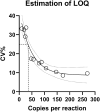
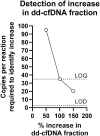
In solid organ transplantation, donor-derived cell-free DNA (dd-cfDNA) is a promising universal noninvasive biomarker for allograft health, where high levels of dd-cfDNA indicate organ damage. Using Droplet Digital PCR (ddPCR), we aimed to develop an assay setup for monitoring organ health. We aimed to identify the least distinguishable percentage-point increase in the fraction of minute amounts of cfDNA in a large cfDNA background by using assays targeting single nucleotide polymorphisms (SNPs). We mimicked a clinical sample from a recipient in a number of spike-in experiments, where cfDNA from healthy volunteers were mixed. A total of 40 assays were tested and approved by qPCR and ddPCR. Limit of detection (LOD) was demonstrated to be approximately 3 copies per reaction, observed at a fraction of 0.002%, and which would equal 6 copies per mL plasma. Limit of quantification (LOQ) was 35 copies per reaction, estimated to 0.038%. The lowest detectable increase in percentage point of dd-cfDNA was approximately 0.04%. Our results demonstrated that ddPCR has great sensitivity, high precision, and exceptional ability to quantify low levels of cfDNA. The ability to distinguish small differences in mimicking dd-cfDNA was far beyond the desired capability. While these methodological data are promising, further prospective studies are needed to determine the clinical utility of the proposed method.
Copyright: © 2023 Clausen et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

