EndophilinA-dependent coupling between activity-induced calcium influx and synaptic autophagy is disrupted by a Parkinson-risk mutation
- PMID: 36827984
- PMCID: PMC10166451
- DOI: 10.1016/j.neuron.2023.02.001
EndophilinA-dependent coupling between activity-induced calcium influx and synaptic autophagy is disrupted by a Parkinson-risk mutation
Abstract
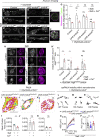
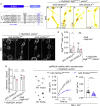
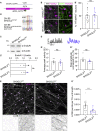
Neuronal activity causes use-dependent decline in protein function. However, it is unclear how this is coupled to local quality control mechanisms. We show in Drosophila that the endocytic protein Endophilin-A (EndoA) connects activity-induced calcium influx to synaptic autophagy and neuronal survival in a Parkinson disease-relevant fashion. Mutations in the disordered loop, including a Parkinson disease-risk mutation, render EndoA insensitive to neuronal stimulation and affect protein dynamics: when EndoA is more flexible, its mobility in membrane nanodomains increases, making it available for autophagosome formation. Conversely, when EndoA is more rigid, its mobility reduces, blocking stimulation-induced autophagy. Balanced stimulation-induced autophagy is required for dopagminergic neuron survival, and a variant in the human ENDOA1 disordered loop conferring risk to Parkinson disease also blocks nanodomain protein mobility and autophagy both in vivo and in human-induced dopaminergic neurons. Thus, we reveal a mechanism that neurons use to connect neuronal activity to local autophagy and that is critical for neuronal survival.
Keywords: Ca(2+) influx; Parkinson disease; endophilinA; neuronal activity; synaptic autophagy.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests P.V. is the scientific founder of Jay Therapeutics. All other authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

