Large-Scale Immunopeptidome Analysis Reveals Recurrent Posttranslational Splicing of Cancer- and Immune-Associated Genes
- PMID: 36828127
- PMCID: PMC10119686
- DOI: 10.1016/j.mcpro.2023.100519
Large-Scale Immunopeptidome Analysis Reveals Recurrent Posttranslational Splicing of Cancer- and Immune-Associated Genes
Abstract
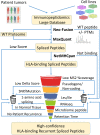
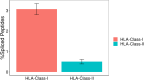
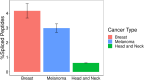
Posttranslational spliced peptides (PTSPs) are a unique class of peptides that have been found to be presented by HLA class-I molecules in cancer. Thus far, no consensus has been reached on the proportion of PTSPs in the immunopeptidome, with estimates ranging from 2% to as high as 45% and stirring significant debate. Furthermore, the role of the HLA class-II pathway in PTSP presentation has been studied only in diabetes. Here, we exploit our large-scale cancer peptidomics database and our newly devised pipeline for filtering spliced peptide predictions to identify recurring spliced peptides, both for HLA class-I and class-II complexes. Our results indicate that HLA class-I-spliced peptides account for a low percentage of the immunopeptidome (less than 3.1%) yet are larger in number relative to other types of identified aberrant peptides. Therefore, spliced peptides significantly contribute to the repertoire of presented peptides in cancer cells. In addition, we identified HLA class-II-bound spliced peptides, but to a lower extent (less than 0.5%). The identified spliced peptides include cancer- and immune-associated genes, such as the MITF oncogene, DAPK1 tumor suppressor, and HLA-E, which were validated using synthetic peptides. The potential immunogenicity of the DAPK1- and HLA-E-derived PTSPs was also confirmed. In addition, a reanalysis of our published mouse single-cell clone immunopeptidome dataset showed that most of the spliced peptides were found repeatedly in a large number of the single-cell clones. Establishing a novel search-scheme for the discovery and evaluation of recurring PTSPs among cancer patients may assist in identifying potential novel targets for immunotherapy.
Keywords: HLA class-I; HLA class-II; immunopeptidomics; neoantigens; posttranslational spliced peptides.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of the article.
Figures






References
-
- Faridi P., Woods K., Ostrouska S., Deceneux C., Aranha R., Duscharla D., et al. Spliced peptides and cytokine-driven changes in the immunopeptidome of melanoma. Cancer Immunol. Res. 2020;8:1322–1334. - PubMed
-
- Bartok O., Pataskar A., Nagel R., Laos M., Goldfarb E., Hayoun D., et al. Anti-tumour immunity induces aberrant peptide presentation in melanoma. Nature. 2021;590:332–337. - PubMed
-
- Vigneron N., Stroobant V., Chapiro J., Ooms A., Degiovanni G., Morel S., et al. An antigenic peptide produced by peptide splicing in the proteasome. Science. 2004;304:587–590. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

