Metabolome-Based Classification of Snake Venoms by Bioinformatic Tools
- PMID: 36828475
- PMCID: PMC9963137
- DOI: 10.3390/toxins15020161
Metabolome-Based Classification of Snake Venoms by Bioinformatic Tools
Abstract
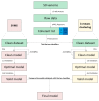
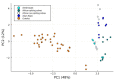
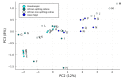
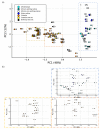
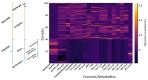
Snakebite is considered a neglected tropical disease, and it is one of the most intricate ones. The variability found in snake venom is what makes it immensely complex to study. These variations are present both in the big and the small molecules found in snake venom. This study focused on examining the variability found in the venom's small molecules (i.e., mass range of 100-1000 Da) between two main families of venomous snakes-Elapidae and Viperidae-managing to create a model able to classify unknown samples by means of specific features, which can be extracted from their LC-MS data and output in a comprehensive list. The developed model also allowed further insight into the composition of snake venom by highlighting the most relevant metabolites of each group by clustering similarly composed venoms. The model was created by means of support vector machines and used 20 features, which were merged into 10 principal components. All samples from the first and second validation data subsets were correctly classified. Biological hypotheses relevant to the variation regarding the metabolites that were identified are also given.
Keywords: data analysis; metabolomics; script-controlled peak integration; venom variation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Mohapatra B., Warrell D.A., Suraweera W., Bhatia P., Dhingra N., Jotkar R.M., Rodriguez P.S., Mishra K., Whitaker R., Jha P., et al. Snakebite Mortality in India: A Nationally Representative Mortality Survey. PLoS Negl. Trop. Dis. 2011;5:e1018. doi: 10.1371/journal.pntd.0001018. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

