S100 family proteins are linked to organoid morphology and EMT in pancreatic cancer
- PMID: 36828915
- PMCID: PMC10154348
- DOI: 10.1038/s41418-023-01126-z
S100 family proteins are linked to organoid morphology and EMT in pancreatic cancer
Erratum in
-
Correction to: S100 family proteins are linked to organoid morphology and EMT in pancreatic cancer.Cell Death Differ. 2023 May;30(5):1400. doi: 10.1038/s41418-023-01151-y. Cell Death Differ. 2023. PMID: 36949280 Free PMC article. No abstract available.
Abstract
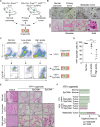
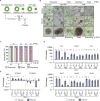
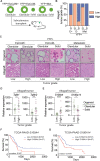
Epithelial-mesenchymal transition (EMT) is a continuum that includes epithelial, partial EMT, and mesenchymal states, each of which is associated with cancer progression, invasive capabilities, and ultimately, metastasis. We used a lineage-traced sporadic model of pancreatic cancer to generate a murine organoid biobank from primary and secondary tumors, including sublines that underwent partial EMT and complete EMT. Using an unbiased proteomics approach, we found that organoid morphology predicts the EMT state, and the solid organoids are associated with a partial EMT signature. We also observed that exogenous TGFβ1 induces solid organoid morphology that is associated with changes in the S100 family, complete EMT, and the formation of high-grade tumors. S100A4 may be a useful biomarker for predicting EMT state, disease progression, and outcome in patients with pancreatic cancer.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest. TLP has consulted for enterprises involved in biological drug development (Mestag Therapeutics, Enleofen Ltd). MDWG has consulted for enterprises involved in biological drug development (Mestag Therapeutics).
Figures



References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA: A Cancer J Clin. 2020;70:7–30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

