Immunoinformatics-aided design of a new multi-epitope vaccine adjuvanted with domain 4 of pneumolysin against Streptococcus pneumoniae strains
- PMID: 36829109
- PMCID: PMC9951839
- DOI: 10.1186/s12859-023-05175-6
Immunoinformatics-aided design of a new multi-epitope vaccine adjuvanted with domain 4 of pneumolysin against Streptococcus pneumoniae strains
Abstract
Background: Streptococcus pneumoniae (Pneumococcus) has remained a leading cause of fatal infections such as pneumonia, meningitis, and sepsis. Moreover, this pathogen plays a major role in bacterial co-infection in patients with life-threatening respiratory virus diseases such as influenza and COVID-19. High morbidity and mortality in over one million cases, especially in very young children and the elderly, are the main motivations for pneumococcal vaccine development. Due to the limitations of the currently marketed polysaccharide-based vaccines, non-serotype-specific protein-based vaccines have received wide research interest in recent years. One step further is to identify high antigenic regions within multiple highly-conserved proteins in order to develop peptide vaccines that can affect various stages of pneumococcal infection, providing broader serotype coverage and more effective protection. In this study, immunoinformatics tools were used to design an effective multi-epitope vaccine in order to elicit neutralizing antibodies against multiple strains of pneumococcus.
Results: The B- and T-cell epitopes from highly protective antigens PspA (clades 1-5) and PhtD were predicted and immunodominant peptides were linked to each other with proper linkers. The domain 4 of Ply, as a potential TLR4 agonist adjuvant candidate, was attached to the end of the construct to enhance the immunogenicity of the epitope vaccine. The evaluation of the physicochemical and immunological properties showed that the final construct was stable, soluble, antigenic, and non-allergenic. Furthermore, the protein was found to be acidic and hydrophilic in nature. The protein 3D-structure was built and refined, and the Ramachandran plot, ProSA-web, ERRAT, and Verify3D validated the quality of the final model. Molecular docking analysis showed that the designed construct via Ply domain 4 had a strong interaction with TLR4. The structural stability of the docked complex was confirmed by molecular dynamics. Finally, codon optimization was performed for gene expression in E. coli, followed by in silico cloning in the pET28a(+) vector.
Conclusion: The computational analysis of the construct showed acceptable results, however, the suggested vaccine needs to be experimentally verified in laboratory to ensure its safety and immunogenicity.
Keywords: Domain 4 of pneumolysin (Ply4); Immunoinformatics; Multi-epitope vaccine; Pneumococcal histidine triad protein D (PhtD); Pneumococcal surface protein A (PspA); Protein TLR agonist adjuvant.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



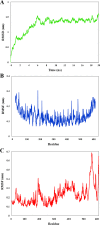

References
-
- Abdollahi S, Siadat SD, Shapouri R, Mirzaei B, Mousavi SF, Nikbin VS, Moosavi SH. Antibiotic susceptibility and prevalence of adhesion genes in Streptococcus pneumoniae isolates detected in carrier children in Tehran. Jundishapur J Microbiol. 2018;11:6. doi: 10.5812/jjm.13256. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

