Does multi-way, long-range chromatin contact data advance 3D genome reconstruction?
- PMID: 36829114
- PMCID: PMC9951495
- DOI: 10.1186/s12859-023-05170-x
Does multi-way, long-range chromatin contact data advance 3D genome reconstruction?
Abstract
Background: Methods for inferring the three-dimensional (3D) configuration of chromatin from conformation capture assays that provide strictly pairwise interactions, notably Hi-C, utilize the attendant contact matrix as input. More recent assays, in particular split-pool recognition of interactions by tag extension (SPRITE), capture multi-way interactions instead of solely pairwise contacts. These assays yield contacts that straddle appreciably greater genomic distances than Hi-C, in addition to instances of exceptionally high-order chromatin interaction. Such attributes are anticipated to be consequential with respect to 3D genome reconstruction, a task yet to be undertaken with multi-way contact data. However, performing such 3D reconstruction using distance-based reconstruction techniques requires framing multi-way contacts as (pairwise) distances. Comparing approaches for so doing, and assessing the resultant impact of long-range and multi-way contacts, are the objectives of this study.
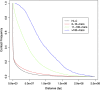
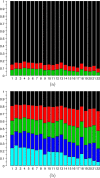
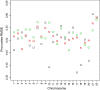
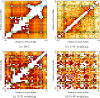
Results: We obtained 3D reconstructions via multi-dimensional scaling under a variety of weighting schemes for mapping SPRITE multi-way contacts to pairwise distances. Resultant configurations were compared following Procrustes alignment and relationships were assessed between associated Procrustes root mean square errors and key features such as the extent of multi-way and/or long-range contacts. We found that these features had surprisingly limited influence on 3D reconstruction, a finding we attribute to their influence being diminished by the preponderance of pairwise contacts.
Conclusion: Distance-based 3D genome reconstruction using SPRITE multi-way contact data is not appreciably affected by the weighting scheme used to convert multi-way interactions to pairwise distances.
Keywords: Conformation capture; Multi-dimensional scaling; Pairwise distance; Procrustes alignment; SPRITE.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Assessing stationary distributions derived from chromatin contact maps.BMC Bioinformatics. 2020 Feb 24;21(1):73. doi: 10.1186/s12859-020-3424-y. BMC Bioinformatics. 2020. PMID: 32093610 Free PMC article.
-
Improved accuracy assessment for 3D genome reconstructions.BMC Bioinformatics. 2018 May 30;19(1):196. doi: 10.1186/s12859-018-2214-2. BMC Bioinformatics. 2018. PMID: 29848293 Free PMC article.
-
Reconstruction of 3D genome architecture via a two-stage algorithm.BMC Bioinformatics. 2015 Nov 9;16:373. doi: 10.1186/s12859-015-0799-2. BMC Bioinformatics. 2015. PMID: 26553003 Free PMC article.
-
Mapping the 3D genome architecture.Comput Struct Biotechnol J. 2024 Dec 23;27:89-101. doi: 10.1016/j.csbj.2024.12.018. eCollection 2025. Comput Struct Biotechnol J. 2024. PMID: 39816913 Free PMC article. Review.
-
SPRITE: a genome-wide method for mapping higher-order 3D interactions in the nucleus using combinatorial split-and-pool barcoding.Nat Protoc. 2022 Jan;17(1):36-75. doi: 10.1038/s41596-021-00633-y. Epub 2022 Jan 10. Nat Protoc. 2022. PMID: 35013617 Free PMC article. Review.
References
-
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range contacts reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. - DOI - PMC - PubMed
-
- Marco A, Meharena HS, Dileep V, Raju RM, Davila-Velderrain J, Zhang AL, Adaikkan C, Young JZ, Gao F, Kellis M, Tsai LH. Mapping the epigenomic and transcriptomic interplay during memory formation and recall in the hippocampal engram ensemble. Nat Neurosci. 2020;23:1606–1617. doi: 10.1038/s41593-020-00717-0. - DOI - PMC - PubMed
-
- Ay F, Bunnik EM, Varoquaux N, Bol SM, Prudhomme J, Vert JP, Noble WS, Le Roch KG. Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Res. 2014;24:974–88. doi: 10.1101/gr.169417.113. - DOI - PMC - PubMed

