Using protein-per-mRNA differences among human tissues in codon optimization
- PMID: 36829202
- PMCID: PMC9951436
- DOI: 10.1186/s13059-023-02868-2
Using protein-per-mRNA differences among human tissues in codon optimization
Abstract
Background: Codon usage and nucleotide composition of coding sequences have profound effects on protein expression. However, while it is recognized that different tissues have distinct tRNA profiles and codon usages in their transcriptomes, the effect of tissue-specific codon optimality on protein synthesis remains elusive.
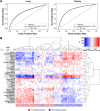
Results: We leverage existing state-of-the-art transcriptomics and proteomics datasets from the GTEx project and the Human Protein Atlas to compute the protein-to-mRNA ratios of 36 human tissues. Using this as a proxy of translational efficiency, we build a machine learning model that identifies codons enriched or depleted in specific tissues. We detect two clusters of tissues with an opposite pattern of codon preferences. We then use these identified patterns for the development of CUSTOM, a codon optimizer algorithm which suggests a synonymous codon design in order to optimize protein production in a tissue-specific manner. In human cell-line models, we provide evidence that codon optimization should take into account particularities of the translational machinery of the tissues in which the target proteins are expressed and that our approach can design genes with tissue-optimized expression profiles.
Conclusions: We provide proof-of-concept evidence that codon preferences exist in tissue-specific protein synthesis and demonstrate its application to synthetic gene design. We show that CUSTOM can be of benefit in biological and biotechnological applications, such as in the design of tissue-targeted therapies and vaccines.
Keywords: Codon optimization; Gene design; Proteomics; Tissue; Transcriptomics; Translation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Watts A, Sankaranarayanan S, Watts A, Raipuria RK. Optimizing protein expression in heterologous system: strategies and tools. Meta Gene. 2021;29:100899. doi: 10.1016/j.mgene.2021.100899. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

