Tenascin C in Lung Diseases
- PMID: 36829478
- PMCID: PMC9953172
- DOI: 10.3390/biology12020199
Tenascin C in Lung Diseases
Abstract
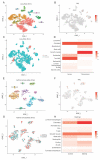
Tenascin C (TNC) is a multifunctional large extracellular matrix protein involved in numerous cellular processes in embryonic development and can be increased in disease, or under conditions of trauma or cell stress in adults. However, the role of TNC in lung diseases remains unclear. In this study, we investigated the expression of TNC during development, in offspring following maternal particulate matter (PM) exposure, asthma, chronic obstructive pulmonary disease (COPD) and lung cancer. TNC expression is increased during lung development in biopsy cells, endothelial cells, mesenchymal cells, and epithelial cells. Maternal PM exposure increased TNC and collagen deposition, which was not affected by the removal of PM exposure after pregnancy. TNC expression was also increased in basal epithelial cells and fibroblasts in patients with asthma and AT2 and endothelial cells in patients with COPD. Furthermore, there was an increase in the expression of TNC in stage II compared to stage IA lung cancer; however, overall survival analysis showed no correlation between levels of TNC and survival. In conclusion, TNC is increased during lung development, in offspring following maternal PM exposure, and in asthma, COPD, and lung cancer tissues. Therefore, targeting TNC may provide a novel therapeutic target for lung diseases.
Keywords: COPD; asthma; foetal programming; lung cancer; lung development; particular matter.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Bradshaw A.D. Principles of Tissue Engineering. Elsevier; Amsterdam, The Netherlands: 2014. Regulation of cell behavior by extracellular proteins; pp. 279–290.
Grants and funding
- APP1158186/National Health and Medical Research Council
- Peter Doherty Fellowship/National Health and Medical Research Council
- CUHK 14212116/Research Grant Council of The Hong Kong Special Administrative Region China
- CRF/C5004-15E/RGC of the Hong Kong Special Administrative Region
- 1-BBW9/the Strategic Focus Area scheme of The Research Institute for Sustainable Urban Development at The Hong Kong Polytechnic University
LinkOut - more resources
Full Text Sources
Miscellaneous

