Methionine Sulfoxide Reductases Suppress the Formation of the [ PSI+] Prion and Protein Aggregation in Yeast
- PMID: 36829961
- PMCID: PMC9952077
- DOI: 10.3390/antiox12020401
Methionine Sulfoxide Reductases Suppress the Formation of the [ PSI+] Prion and Protein Aggregation in Yeast
Abstract
Prions are self-propagating, misfolded forms of proteins associated with various neurodegenerative diseases in mammals and heritable traits in yeast. How prions form spontaneously into infectious amyloid-like structures without underlying genetic changes is poorly understood. Previous studies have suggested that methionine oxidation may underlie the switch from a soluble protein to the prion form. In this current study, we have examined the role of methionine sulfoxide reductases (MXRs) in protecting against de novo formation of the yeast [PSI+] prion, which is the amyloid form of the Sup35 translation termination factor. We show that [PSI+] formation is increased during normal and oxidative stress conditions in mutants lacking either one of the yeast MXRs (Mxr1, Mxr2), which protect against methionine oxidation by reducing the two epimers of methionine-S-sulfoxide. We have identified a methionine residue (Met124) in Sup35 that is important for prion formation, confirming that direct Sup35 oxidation causes [PSI+] prion formation. [PSI+] formation was less pronounced in mutants simultaneously lacking both MXR isoenzymes, and we show that the morphology and biophysical properties of protein aggregates are altered in this mutant. Taken together, our data indicate that methionine oxidation triggers spontaneous [PSI+] prion formation, which can be alleviated by methionine sulfoxide reductases.
Keywords: methionine oxidation; methionine sulfoxide reductase; oxidative stress; prions; protein aggregation; yeast.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


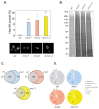
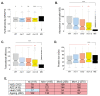

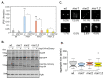
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

