Comparative Genomics Identifies Novel Genetic Changes Associated with Oxacillin, Vancomycin and Daptomycin Susceptibility in ST100 Methicillin-Resistant Staphylococcus aureus
- PMID: 36830286
- PMCID: PMC9952151
- DOI: 10.3390/antibiotics12020372
Comparative Genomics Identifies Novel Genetic Changes Associated with Oxacillin, Vancomycin and Daptomycin Susceptibility in ST100 Methicillin-Resistant Staphylococcus aureus
Abstract
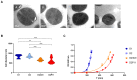
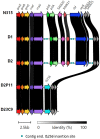
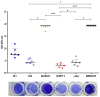
Infections due to vancomycin-intermediate S. aureus (VISA) and heterogeneous VISA (hVISA) represent a serious concern due to their association with vancomycin treatment failure. However, the underlying molecular mechanism responsible for the hVISA/VISA phenotype is complex and not yet fully understood. We have previously characterized two ST100-MRSA-hVISA clinical isolates recovered before and after 40 days of vancomycin treatment (D1 and D2, respectively) and two in vitro VISA derivatives (D23C9 and D2P11), selected independently from D2 in the presence of vancomycin. This follow-up study was aimed at further characterizing these isogenic strains and obtaining their whole genome sequences to unravel changes associated with antibiotic resistance. It is interesting to note that none of these isogenic strains carry SNPs in the regulatory operons vraUTSR, walKR and/or graXRS. Nonetheless, genetic changes including SNPs, INDELs and IS256 genomic insertions/rearrangements were found both in in vivo and in vitro vancomycin-selected strains. Some were found in the downstream target genes of the aforementioned regulatory operons, which are involved in cell wall and phosphate metabolism, staphylococcal growth and biofilm formation. Some of the genetic changes reported herein have not been previously associated with vancomycin, daptomycin and/or oxacillin resistance in S. aureus.
Keywords: IS256; MRSA; S. aureus; ST100; WGS; hVISA/VISA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- WHO List of Bacteria for Which New Antibiotics Are Urgently Needed. [(accessed on 20 December 2022)]. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-....
-
- Shariati A., Dadashi M., Moghadam M.T., van Belkum A., Yaslianifard S., Darban-Sarokhalil D. Global Prevalence and Distribution of Vancomycin Resistant, Vancomycin Intermediate and Heterogeneously Vancomycin Intermediate Staphylococcus Aureus Clinical Isolates: A Systematic Review and Meta-Analysis. Sci. Rep. 2020;10:12689. doi: 10.1038/s41598-020-69058-z. - DOI - PMC - PubMed
-
- Gostev V.V., Popenko L.N., Chernen’kaia T.V., Naumenko Z.S., Voroshilova T.M., Zakharova I.A., Khokhlova O.E., Kruglov A.N., Ershova M.G., Angelova S.N., et al. Estimation of MRSA susceptibility to oxacillin, cefoxitine, vancomycin and daptomycin. Antibiot. Chemoterapy. 2013;58:13–20. - PubMed
-
- Di Gregorio S., Perazzi B., Ordoñez A.M., De Gregorio S., Foccoli M., Lasala M.B., García S., Vay C., Famiglietti A., Mollerach M. Clinical, Microbiological, and Genetic Characteristics of Heteroresistant Vancomycin-Intermediate Staphylococcus Aureus Bacteremia in a Teaching Hospital. Microb. Drug Resist. 2015;21:25–34. doi: 10.1089/mdr.2014.0190. - DOI - PMC - PubMed
Grants and funding
- UBACYT 2018-2020-20020170100665BA/University of Buenos Aires
- PIP 2015 11220150100694CO/National Scientific and Technical Research Council
- Préstamo BID PICT-2016-1726/Agencia Nacional de Promoción Científica y Tecnológica
- Préstamo BID PICT-2018-03068/Agencia Nacional de Promoción Científica y Tecnológica
LinkOut - more resources
Full Text Sources
Miscellaneous

