Growth Stages and Inter-Species Gut Microbiota Composition and Function in Captive Red Deer (Cervus elaphus alxaicus) and Blue Sheep (Pseudois nayaur)
- PMID: 36830340
- PMCID: PMC9951700
- DOI: 10.3390/ani13040553
Growth Stages and Inter-Species Gut Microbiota Composition and Function in Captive Red Deer (Cervus elaphus alxaicus) and Blue Sheep (Pseudois nayaur)
Abstract
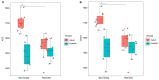
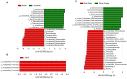
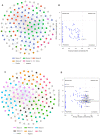
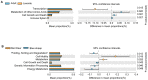
Blue sheep and red deer, second-class key protected animals in China, are sympatric species with a high degree of overlap of food resources in the Helan Mountains, China. Previous studies with blue sheep and red deer in nature have shown that their physiology is closely related to their gut microbiota. However, growth stages and changes occurring in these species in captivity are still unknown. Thus, 16S rRNA gene sequencing was used to explore diversity, composition and function of the gut microbiota in these two animal species. The diversity and structure of the gut microbiota in captive blue sheep and red deer changed at different growth stages, but the dominant microbiota phyla in the gut microbiota remained stable, which was composed of the phyla Firmicutes, Bacteroidetes and Verrucomicrobia. Moreover, gut microbiota diversity in juvenile blue sheep and red deer was low, with the potential for further colonization. Functional predictions showed differences such as red deer transcription being enriched in adults, and blue sheep adults having a higher cell wall/membrane/envelope biogenesis than juveniles. Microbial changes between blue sheep and red deer at different growth stages and between species mainly depend on the abundance of the microbiota, rather than the increase and absence of the bacterial taxa.
Keywords: 16S rRNA gene sequencing; blue sheep; captivity; growth stage; gut microbes; red deer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Wang B., Liu X., Bian Y., Sun G., Wang H., Zhang J., Zhang Z., Zou X. Correlation analysis of breast fibroadenoma and the intestinal flora based on 16S rRNA sequencing. Oncol. Transl. Med. 2021;7:269–274.
-
- Franza L., Carusi V., Altamura S., Gasbarrini A., Caraffa A., Kritas S.K., Ronconi G., Gallenga C.E., Di Virgilio F., Pandolfi F. Gut microbiota and immunity in common variable immunodeficiency: Crosstalk with pro-inflammatory cytokines. J. Biol. Regul. Homeost. Agents. 2019;33:315–319. - PubMed
-
- Shipo L., Xing H., Liyun G., Li S.P., Heng X., Guo L.Y., Lessing D.J., Chu W.H. SCFAs improve disease resistance via modulate gut microbiota, enhance immune response and increase antioxidative capacity in the host. Fish Shellfish Immunol. 2022;120:560–568. - PubMed
-
- Tsegay T., Xuexiang W., Qiang H., Qianwen D., Hongliang L., Chao R., Yalin Y., Liqiu X., Shaojun W., Zhigang Z., et al. Dietary SWF® enhanced growth performance and disease resistance in hybrid sturgeon (Acipenser baerii × Acipenser schrenckii) mediated by the gut microbiota. Aquac. Rep. 2020;17:100346.
Grants and funding
- (31870512, 32071649 and 32070519)/National Natural Science Foundation of China
- (NLF2020-93-1)/Ningxia Helan Mountain National Nature Reserve Administration
- no number/Heilongjiang Touyan Innovation Team Program for Forest Ecology and Conservation
- 2020BEG02001/The Key Research and Development Program of Ningxia Hui Autonomous Region, China
LinkOut - more resources
Full Text Sources

