Hepatic Transcriptome Analysis Reveals Genes, Polymorphisms, and Molecules Related to Lamb Tenderness
- PMID: 36830461
- PMCID: PMC9951696
- DOI: 10.3390/ani13040674
Hepatic Transcriptome Analysis Reveals Genes, Polymorphisms, and Molecules Related to Lamb Tenderness
Abstract
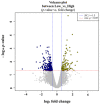
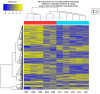
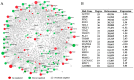
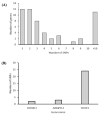
Tenderness is a key meat quality trait that determines the public acceptance of lamb consumption, so genetic improvement toward lamb with higher tenderness is pivotal for a sustainable sheep industry. However, unravelling the genomics controlling the tenderness is the first step. Therefore, this study aimed to identify the transcriptome signatures and polymorphisms related to divergent lamb tenderness using RNA deep sequencing. Since the molecules and enzymes that control muscle growth and tenderness are metabolized and synthesized in the liver, hepatic tissues of ten sheep with divergent phenotypes: five high- and five low-lamb tenderness samples were applied for deep sequencing. Sequence analysis identified the number of reads ranged from 21.37 to 25.37 million bases with a mean value of 22.90 million bases. In total, 328 genes are detected as differentially expressed (DEGs) including 110 and 218 genes that were up- and down-regulated, respectively. Pathway analysis showed steroid hormone biosynthesis as the dominant pathway behind the lamb tenderness. Gene expression analysis identified the top high (such as TP53INP1, CYP2E1, HSD17B13, ADH1C, and LPIN1) and low (such as ANGPTL2, IGFBP7, FABP5, OLFML3, and THOC5) expressed candidate genes. Polymorphism and association analysis revealed that mutation in OLFML3, ANGPTL2, and THOC5 genes could be potential candidate markers for tenderness in sheep. The genes and pathways identified in this study cause variation in tenderness, thus could be potential genetic markers to improve meat quality in sheep. However, further validation is needed to confirm the effect of these markers in different sheep populations so that these could be used in a selection program for lamb with high tenderness.
Keywords: genetic marker; meat quality; next generation genome sequencing; sheep; single nucleotide polymorphism.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




Similar articles
-
Hepatic transcriptome analysis identifies genes, polymorphisms and pathways involved in the fatty acids metabolism in sheep.PLoS One. 2021 Dec 23;16(12):e0260514. doi: 10.1371/journal.pone.0260514. eCollection 2021. PLoS One. 2021. PMID: 34941886 Free PMC article.
-
A comprehensive study of CYP2E1 and its role in carcass characteristics and chemical lamb meat quality in different Indonesian sheep breeds.PLoS One. 2024 Sep 9;19(9):e0310336. doi: 10.1371/journal.pone.0310336. eCollection 2024. PLoS One. 2024. PMID: 39250496 Free PMC article.
-
Transcriptome signature of liver tissue with divergent mutton odour and flavour using RNA deep sequencing.Gene. 2018 Nov 15;676:86-94. doi: 10.1016/j.gene.2018.06.086. Epub 2018 Jun 27. Gene. 2018. PMID: 29958950
-
Transcriptome analysis reveals candidate genes of the synthesis of branched-chain fatty acids related to mutton flavor in the lamb liver using Allium mongolicum Regel extract.J Anim Sci. 2022 Sep 1;100(9):skac256. doi: 10.1093/jas/skac256. J Anim Sci. 2022. PMID: 35946924 Free PMC article.
-
Effect of Pre-Slaughter Practises and Early Post-Mortem Interventions on Sheep Meat Tenderness and Its Impact on Microbial Status.Foods. 2022 Jan 11;11(2):181. doi: 10.3390/foods11020181. Foods. 2022. PMID: 35053913 Free PMC article. Review.
Cited by
-
Comparative transcriptome analysis reveals genes associated with variation in liver copper concentration in Polish Merino sheep.Sci Rep. 2025 Jun 4;15(1):19576. doi: 10.1038/s41598-025-04330-8. Sci Rep. 2025. PMID: 40467731 Free PMC article.
-
Integrative Analysis of Transcriptomics and Metabolomics Provides Insights into Meat Quality Differences in Hu Sheep with Different Carcass Performance.Foods. 2025 Jul 15;14(14):2477. doi: 10.3390/foods14142477. Foods. 2025. PMID: 40724298 Free PMC article.
-
Effect of supplementing lysins and methionine to low-protein diets on growth performance, hepatic antioxidant capacity, immune status, and glycolytic activity of tibetan sheep.BMC Genomics. 2024 Jun 4;25(1):557. doi: 10.1186/s12864-024-10480-2. BMC Genomics. 2024. PMID: 38834972 Free PMC article.
-
Potential candidate genes influencing meat production phenotypic traits in sheep: a review.Front Vet Sci. 2025 Jul 16;12:1616533. doi: 10.3389/fvets.2025.1616533. eCollection 2025. Front Vet Sci. 2025. PMID: 40740300 Free PMC article. Review.
-
IGFBP7 promotes the proliferation and differentiation of primary myoblasts and intramuscular preadipocytes in chicken.Poult Sci. 2024 Dec;103(12):104258. doi: 10.1016/j.psj.2024.104258. Epub 2024 Sep 2. Poult Sci. 2024. PMID: 39293261 Free PMC article.
References
-
- FAO (Food and Agriculture Organization) Agriculture and Consumer Protection Department Animal Production and Health: Meat Quality. 2014. [(accessed on 1 February 2021)]. Available online: http://www.fao.org/ag/againfo/themes/en/meat/quality_meat.html.
-
- Thu D.T.N. Meat quality: Understanding of meat tenderness and influence of fat content on meat flavor. Sci. Technol. Dev. 2006;9:65–70.
-
- Shackelford S.D., Morgan J.B., Cross H.R., Savell J.W. Identification of threshold levels for warner-bratzler shear force in beef top loin steaks. J. Muscle Foods. 1991;2:289–296. doi: 10.1111/j.1745-4573.1991.tb00461.x. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

