Comparative Research: Regulatory Mechanisms of Ribosomal Gene Transcription in Saccharomyces cerevisiae and Schizosaccharomyces pombe
- PMID: 36830657
- PMCID: PMC9952952
- DOI: 10.3390/biom13020288
Comparative Research: Regulatory Mechanisms of Ribosomal Gene Transcription in Saccharomyces cerevisiae and Schizosaccharomyces pombe
Abstract
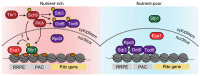
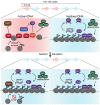
Restricting ribosome biosynthesis and assembly in response to nutrient starvation is a universal phenomenon that enables cells to survive with limited intracellular resources. When cells experience starvation, nutrient signaling pathways, such as the target of rapamycin (TOR) and protein kinase A (PKA), become quiescent, leading to several transcription factors and histone modification enzymes cooperatively and rapidly repressing ribosomal genes. Fission yeast has factors for heterochromatin formation similar to mammalian cells, such as H3K9 methyltransferase and HP1 protein, which are absent in budding yeast. However, limited studies on heterochromatinization in ribosomal genes have been conducted on fission yeast. Herein, we shed light on and compare the regulatory mechanisms of ribosomal gene transcription in two species with the latest insights.
Keywords: TOR pathway; TORC1; budding yeast; epigenetics; fission yeast; gene regulation; heterochromatin; ribosome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

