In Vivo Protein-Protein Binding Competition Assay Based on Split-GFP Reassembly: Proof of Concept
- PMID: 36830723
- PMCID: PMC9952896
- DOI: 10.3390/biom13020354
In Vivo Protein-Protein Binding Competition Assay Based on Split-GFP Reassembly: Proof of Concept
Abstract
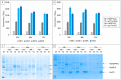
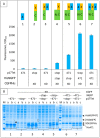
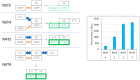
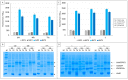
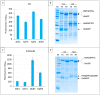
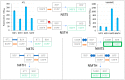
The split-green fluorescent protein (GFP) reassembly assay is a well-established approach to study protein-protein interactions (PPIs). In this assay, when two interacting proteins X and Y, respectively fused to residues 1-157 and to residues 158-237 of GFP, are co-expressed in E. coli, the two GFP halves are brought to sufficient proximity to reassociate and fold to recreate the functional GFP. At constant protein expression level, the intensity of fluorescence produced by the bacteria is proportional to the binding affinity of X to Y. We hypothesized that adding a third partner (Z) endowed with an affinity for either X or Y would lead to an in vivo competition assay. We report here the different steps of the set-up of this competition assay, and define the experimental conditions required to obtained reliable results. Results show that this competition assay is a potentially interesting tool for screening libraries of binding inhibitors, Z being either a protein or a chemical reagent.
Keywords: binding competition assays; fluorescence; intrinsically disordered proteins; protein complementation assays; protein–protein interactions.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

