Alterations in Gut Microbiota Composition in Patients with COVID-19: A Pilot Study of Whole Hypervariable 16S rRNA Gene Sequencing
- PMID: 36830905
- PMCID: PMC9953267
- DOI: 10.3390/biomedicines11020367
Alterations in Gut Microbiota Composition in Patients with COVID-19: A Pilot Study of Whole Hypervariable 16S rRNA Gene Sequencing
Abstract
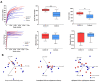
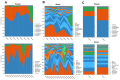
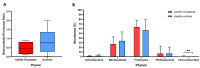
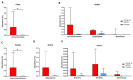
It is crucial to consider the importance of the microbiome and the gut-lung axis in the context of SARS-CoV-2 infection. This pilot study examined the fecal microbial composition of patients with COVID-19 following a 3-month recovery. Using for the first time metagenomic analysis based on all hypervariable regions (V1-V9) of the 16S rRNA gene, we have identified 561 microbial species; however, 17 were specific only for the COVID-19 group (n = 8). The patients' cohorts revealed significantly greater alpha diversity of the gut microbiota compared to healthy controls (n = 14). This finding has been demonstrated by operational taxonomic units (OTUs) richness (p < 0.001) and Chao1 index (p < 0.01). The abundance of the phylum Verrucomicrobia was 30 times higher in COVID-19 patients compared to healthy subjects. Accordingly, this disproportion was also noted at other taxonomic levels: in the class Verrucomicrobiae, the family Verrucomicrobiaceae, and the genus Akkermansia. Elevated pathobionts such as Escherichia coli, Bilophila wadsworthia, and Parabacteroides distasonis were found in COVID-19 patients. Considering the gut microbiota's ability to disturb the immune response, our findings suggest the importance of the enteric microbiota in the course of SARS-CoV-2 infection. This pilot study shows that the composition of the microbial community may not be fully restored in individuals with SARS-CoV-2 following a 3-month recovery.
Keywords: COVID-19; dysbiosis; gastrointestinal tract; gut microbiome; gut microbiota; gut–lung axis; microbiota; recovered COVID-19 patient.
Conflict of interest statement
Authors declare no conflict of interest.
Figures


References
-
- WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. [(accessed on 16 December 2021)]. Available online: https://www.who.int/director-general/speeches/detail/who-director-genera....
-
- Sharma A., Bhatt N.S., St Martin A., Abid M.B., Bloomquist J., Chemaly R.F., Dandoy C., Gauthier J., Gowda L., Perales M.-A., et al. Clinical Characteristics and Outcomes of COVID-19 in Haematopoietic Stem-Cell Transplantation Recipients: An Observational Cohort Study. Lancet Haematol. 2021;8:e185–e193. doi: 10.1016/S2352-3026(20)30429-4. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

