DNA Gyrase as a Target for Quinolones
- PMID: 36830908
- PMCID: PMC9953508
- DOI: 10.3390/biomedicines11020371
DNA Gyrase as a Target for Quinolones
Abstract
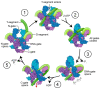
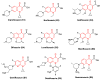
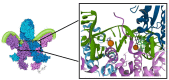
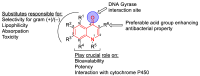
Bacterial DNA gyrase is a type II topoisomerase that can introduce negative supercoils to DNA substrates and is a clinically-relevant target for the development of new antibacterials. DNA gyrase is one of the primary targets of quinolones, broad-spectrum antibacterial agents and are used as a first-line drug for various types of infections. However, currently used quinolones are becoming less effective due to drug resistance. Common resistance comes in the form of mutation in enzyme targets, with this type being the most clinically relevant. Additional mechanisms, conducive to quinolone resistance, are arbitrated by chromosomal mutations and/or plasmid-gene uptake that can alter quinolone cellular concentration and interaction with the target, or affect drug metabolism. Significant synthetic strategies have been employed to modify the quinolone scaffold and/or develop novel quinolones to overcome the resistance problem. This review discusses the development of quinolone antibiotics targeting DNA gyrase to overcome bacterial resistance and reduce toxicity. Moreover, structural activity relationship (SAR) data included in this review could be useful for the development of future generations of quinolone antibiotics.
Keywords: DNA gyrase; drug development; drug-resistance; molecular docking; quinolones.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- O’Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Government of the United Kingdom; London, UK: 2016. Review on Antimicrobial Resistance.
-
- CDC . COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022. U.S. Department of Health and Human Services, CDC; Atlanta, GA, USA: 2022.
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

