Pan-Genomic Regulation of Gene Expression in Normal and Pathological Human Placentas
- PMID: 36831244
- PMCID: PMC9954093
- DOI: 10.3390/cells12040578
Pan-Genomic Regulation of Gene Expression in Normal and Pathological Human Placentas
Abstract
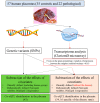
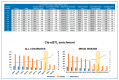
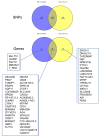
In this study, we attempted to find genetic variants affecting gene expression (eQTL = expression Quantitative Trait Loci) in the human placenta in normal and pathological situations. The analysis of gene expression in placental diseases (Pre-eclampsia and Intra-Uterine Growth Restriction) is hindered by the fact that diseased placental tissue samples are generally taken at earlier gestations compared to control samples. The difference in gestational age is considered a major confounding factor in the transcriptome regulation of the placenta. To alleviate this significant problem, we propose here a novel approach to pinpoint disease-specific cis-eQTLs. By statistical correction for gestational age at sampling as well as other confounding/surrogate variables systematically searched and identified, we found 43 e-genes for which proximal SNPs influence expression level. Then, we performed the analysis again, removing the disease status from the covariates, and we identified 54 e-genes, 16 of which are identified de novo and, thus, possibly related to placental disease. We found a highly significant overlap with previous studies for the list of 43 e-genes, validating our methodology and findings. Among the 16 disease-specific e-genes, several are intrinsic to trophoblast biology and, therefore, constitute novel targets of interest to better characterize placental pathology and its varied clinical consequences. The approach that we used may also be applied to the study of other human diseases where confounding factors have hampered a better understanding of the pathology.
Keywords: expression Quantitative Trait Loci; placenta; preeclampsia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

