In Silico Identification and Functional Characterization of Genetic Variations across DLBCL Cell Lines
- PMID: 36831263
- PMCID: PMC9954129
- DOI: 10.3390/cells12040596
In Silico Identification and Functional Characterization of Genetic Variations across DLBCL Cell Lines
Abstract
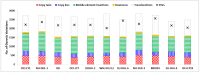
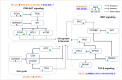
Diffuse large B-cell lymphoma (DLBCL) is the most common form of non-Hodgkin lymphoma and frequently develops through the accumulation of several genetic variations. With the advancement in high-throughput techniques, in addition to mutations and copy number variations, structural variations have gained importance for their role in genome instability leading to tumorigenesis. In this study, in order to understand the genetics of DLBCL pathogenesis, we carried out a whole-genome mutation profile analysis of eleven human cell lines from germinal-center B-cell-like (GCB-7) and activated B-cell-like (ABC-4) subtypes of DLBCL. Analysis of genetic variations including small sequence variants and large structural variations across the cell lines revealed distinct variation profiles indicating the heterogeneous nature of DLBCL and the need for novel patient stratification methods to design potential intervention strategies. Validation and prognostic significance of the variants was assessed using annotations provided for DLBCL samples in cBioPortal for Cancer Genomics. Combining genetic variations revealed new subgroups between the subtypes and associated enriched pathways, viz., PI3K-AKT signaling, cell cycle, TGF-beta signaling, and WNT signaling. Mutation landscape analysis also revealed drug-variant associations and possible effectiveness of known and novel DLBCL treatments. From the whole-genome-based mutation analysis, our findings suggest putative molecular genetics of DLBCL lymphomagenesis and potential genomics-driven precision treatments.
Keywords: cell lines; copy number variations; diffuse large B-cell lymphoma (DLBCL); precision medicine; sequence variations; structural variations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Hans C.P., Weisenburger D.D., Greiner T.C., Gascoyne R.D., Delabie J., Ott G., Müller-Hermelink H.K., Campo E., Braziel R.M., Jaffe E.S., et al. Confirmation of the Molecular Classification of Diffuse Large B-Cell Lymphoma by Immunohistochemistry Using a Tissue Microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. - DOI - PubMed
-
- Choi W.W.L., Weisenburger D.D., Greiner T.C., Piris M.A., Banham A.H., Delabie J., Braziel R.M., Geng H., Iqbal J., Lenz G., et al. A New Immunostain Algorithm Classifies Diffuse Large B-Cell Lymphoma into Molecular Subtypes with High Accuracy. Clin. Cancer Res. 2009;15:5494–5502. doi: 10.1158/1078-0432.CCR-09-0113. - DOI - PMC - PubMed
-
- Carbone P.P., Kaplan H.S., Musshoff K., Smithers D.W., Tubiana M. Report of the Committee on Hodgkin’s Disease Staging Classification. Cancer Res. 1971;31:1860–1861. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

