Decoupling of mRNA and Protein Expression in Aging Brains Reveals the Age-Dependent Adaptation of Specific Gene Subsets
- PMID: 36831282
- PMCID: PMC9954025
- DOI: 10.3390/cells12040615
Decoupling of mRNA and Protein Expression in Aging Brains Reveals the Age-Dependent Adaptation of Specific Gene Subsets
Abstract
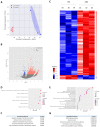
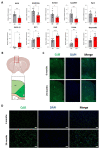
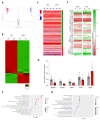
During aging, changes in gene expression are associated with a decline in physical and cognitive abilities. Here, we investigate the connection between changes in mRNA and protein expression in the brain by comparing the transcriptome and proteome of the mouse cortex during aging. Our transcriptomic analysis revealed that aging mainly triggers gene activation in the cortex. We showed that an increase in mRNA expression correlates with protein expression, specifically in the anterior cingulate cortex, where we also observed an increase in cortical thickness during aging. Genes exhibiting an aging-dependent increase of mRNA and protein levels are involved in sensory perception and immune functions. Our proteomic analysis also identified changes in protein abundance in the aging cortex and highlighted a subset of proteins that were differentially enriched but exhibited stable mRNA levels during aging, implying the contribution of aging-related post- transcriptional and post-translational mechanisms. These specific genes were associated with general biological processes such as translation, ribosome assembly and protein degradation, and also important brain functions related to neuroplasticity. By decoupling mRNA and protein expression, we have thus characterized distinct subsets of genes that differentially adjust to cellular aging in the cerebral cortex.
Keywords: aging; brain; cortex; mRNA; protein; proteome; transcriptome.
Conflict of interest statement
The authors have declared no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

